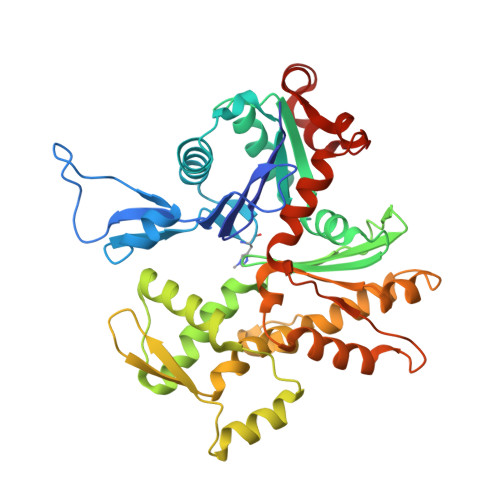
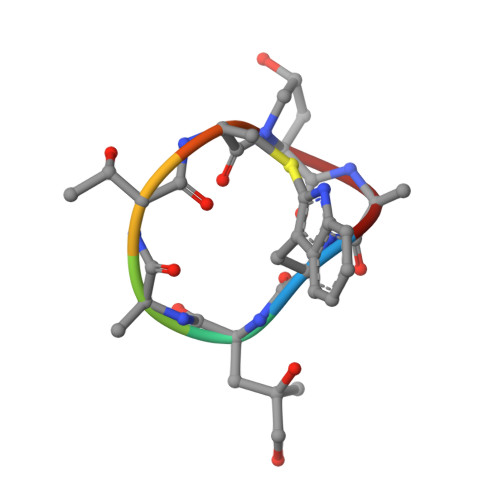
Structural Effects and Functional Implications of Phalloidin and Jasplakinolide Binding to Actin Filaments.
Pospich, S., Merino, F., Raunser, S.(2020) Structure 28: 437-449.e5
- PubMed: 32084355
- DOI: https://doi.org/10.1016/j.str.2020.01.014
- Primary Citation of Related Structures:
6T1Y, 6T20, 6T23, 6T24, 6T25 - PubMed Abstract:
Actin undergoes structural transitions during polymerization, ATP hydrolysis, and subsequent release of inorganic phosphate. Several actin-binding proteins sense specific states during this transition and can thus target different regions of the actin filament. Here, we show in atomic detail that phalloidin, a mushroom toxin that is routinely used to stabilize and label actin filaments, suspends the structural changes in actin, likely influencing its interaction with actin-binding proteins. Furthermore, high-resolution cryoelectron microscopy structures reveal structural rearrangements in F-actin upon inorganic phosphate release in phalloidin-stabilized filaments. We find that the effect of the sponge toxin jasplakinolide differs from the one of phalloidin, despite their overlapping binding site and similar interactions with the actin filament. Analysis of structural conformations of F-actin suggests that stabilizing agents trap states within the natural conformational space of actin.
- Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, Dortmund 44227, Germany.
Organizational Affiliation: