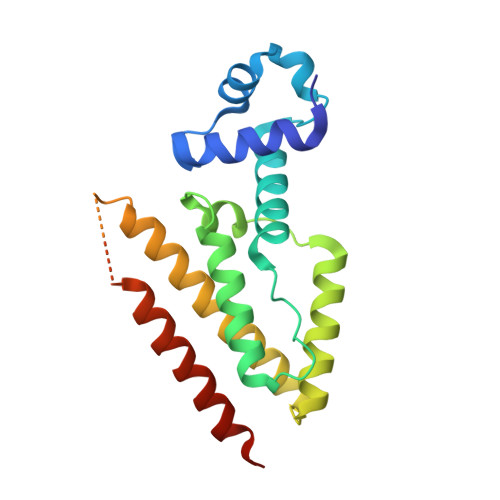
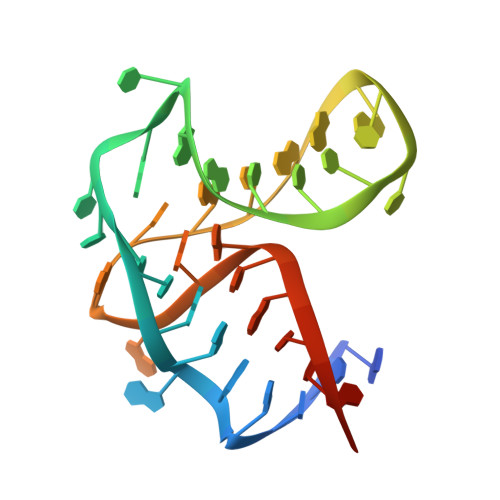
The complex formed between a synthetic RNA aptamer and the transcription repressor TetR is a structural and functional twin of the operator DNA-TetR regulator complex.
Grau, F.C., Jaeger, J., Groher, F., Suess, B., Muller, Y.A.(2020) Nucleic Acids Res 48: 3366-3378
- PubMed: 32052019
- DOI: https://doi.org/10.1093/nar/gkaa083
- Primary Citation of Related Structures:
6SY4, 6SY6 - PubMed Abstract:
RNAs play major roles in the regulation of gene expression. Hence, designer RNA molecules are increasingly explored as regulatory switches in synthetic biology. Among these, the TetR-binding RNA aptamer was selected by its ability to compete with operator DNA for binding to the bacterial repressor TetR. A fortuitous finding was that induction of TetR by tetracycline abolishes both RNA aptamer and operator DNA binding in TetR. This enabled numerous applications exploiting both the specificity of the RNA aptamer and the efficient gene repressor properties of TetR. Here, we present the crystal structure of the TetR-RNA aptamer complex at 2.7 Å resolution together with a comprehensive characterization of the TetR-RNA aptamer versus TetR-operator DNA interaction using site-directed mutagenesis, size exclusion chromatography, electrophoretic mobility shift assays and isothermal titration calorimetry. The fold of the RNA aptamer bears no resemblance to regular B-DNA, and neither does the thermodynamic characterization of the complex formation reaction. Nevertheless, the functional aptamer-binding epitope of TetR is fully contained within its DNA-binding epitope. In the RNA aptamer complex, TetR adopts the well-characterized DNA-binding-competent conformation of TetR, thus revealing how the synthetic TetR-binding aptamer strikes the chords of the bimodal allosteric behaviour of TetR to function as a synthetic regulator.
- Lehrstuhl für Biotechnik, Department of Biology, Friedrich-Alexander University Erlangen-Nuremberg, Henkestr. 91, D-91052 Erlangen, Germany.
Organizational Affiliation: