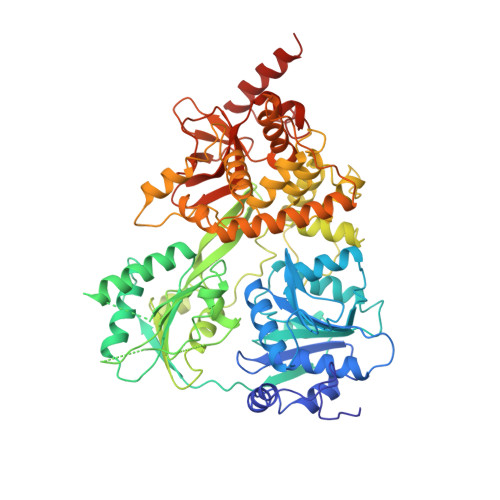
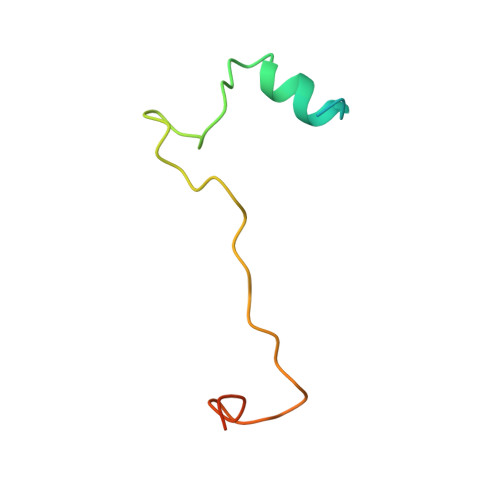
Structural basis for DEAH-helicase activation by G-patch proteins.
Studer, M.K., Ivanovic, L., Weber, M.E., Marti, S., Jonas, S.(2020) Proc Natl Acad Sci U S A 117: 7159-7170
- PubMed: 32179686
- DOI: https://doi.org/10.1073/pnas.1913880117
- Primary Citation of Related Structures:
6SH6, 6SH7 - PubMed Abstract:
RNA helicases of the DEAH/RHA family are involved in many essential cellular processes, such as splicing or ribosome biogenesis, where they remodel large RNA-protein complexes to facilitate transitions to the next intermediate. DEAH helicases couple adenosine triphosphate (ATP) hydrolysis to conformational changes of their catalytic core. This movement results in translocation along RNA, which is held in place by auxiliary C-terminal domains. The activity of DEAH proteins is strongly enhanced by the large and diverse class of G-patch activators. Despite their central roles in RNA metabolism, insight into the molecular basis of G-patch-mediated helicase activation is missing. Here, we have solved the structure of human helicase DHX15/Prp43, which has a dual role in splicing and ribosome assembly, in complex with the G-patch motif of the ribosome biogenesis factor NKRF. The G-patch motif binds in an extended conformation across the helicase surface. It tethers the catalytic core to the flexibly attached C-terminal domains, thereby fixing a conformation that is compatible with RNA binding. Structures in the presence or absence of adenosine diphosphate (ADP) suggest that motions of the catalytic core, which are required for ATP binding, are still permitted. Concomitantly, RNA affinity, helicase, and ATPase activity of DHX15 are increased when G-patch is bound. Mutations that detach one end of the tether but maintain overall binding severely impair this enhancement. Collectively, our data suggest that the G-patch motif acts like a flexible brace between dynamic portions of DHX15 that restricts excessive domain motions but maintains sufficient flexibility for catalysis.
- Institute of Molecular Biology and Biophysics, Department of Biology, Swiss Federal Institute of Technology (ETH) Zürich, 8093 Zürich, Switzerland.
Organizational Affiliation: