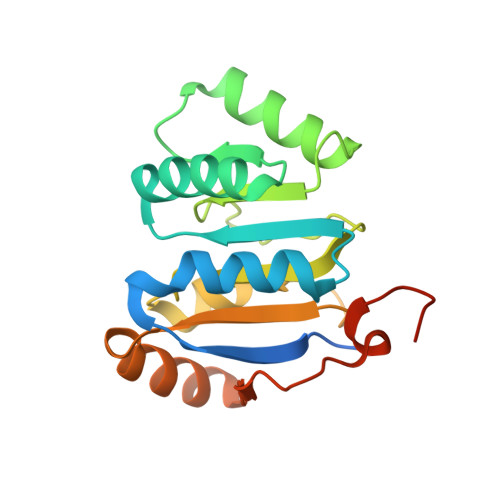
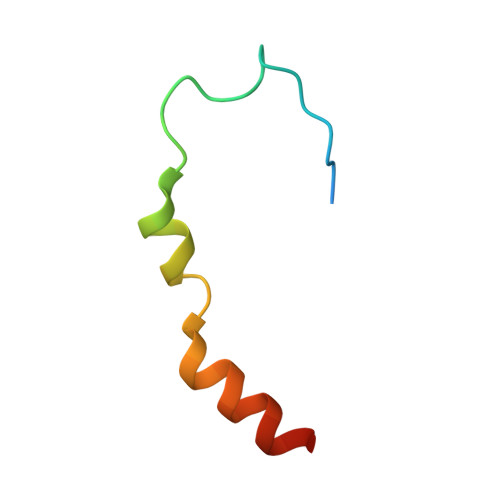
Molecular basis for GIGYF-Me31B complex assembly in 4EHP-mediated translational repression.
Peter, D., Ruscica, V., Bawankar, P., Weber, R., Helms, S., Valkov, E., Igreja, C., Izaurralde, E.(2019) Genes Dev 33: 1355-1360
- PubMed: 31439631
- DOI: https://doi.org/10.1101/gad.329219.119
- Primary Citation of Related Structures:
6S8R, 6S8S - PubMed Abstract:
GIGYF (Grb10-interacting GYF [glycine-tyrosine-phenylalanine domain]) proteins coordinate with 4EHP (eIF4E [eukaryotic initiation factor 4E] homologous protein), the DEAD (Asp-Glu-Ala-Asp)-box helicase Me31B/DDX6, and mRNA-binding proteins to elicit transcript-specific repression. However, the underlying molecular mechanism remains unclear. Here, we report that GIGYF contains a motif necessary and sufficient for direct interaction with Me31B/DDX6. A 2.4 Å crystal structure of the GIGYF-Me31B complex reveals that this motif arranges into a coil connected to a β hairpin on binding to conserved hydrophobic patches on the Me31B RecA2 domain. Structure-guided mutants indicate that 4EHP-GIGYF-DDX6 complex assembly is required for tristetraprolin-mediated down-regulation of an AU-rich mRNA, thus revealing the molecular principles of translational repression.
- Department of Biochemistry, Max Planck Institute for Developmental Biology, D-72076 Tübingen, Germany.
Organizational Affiliation: