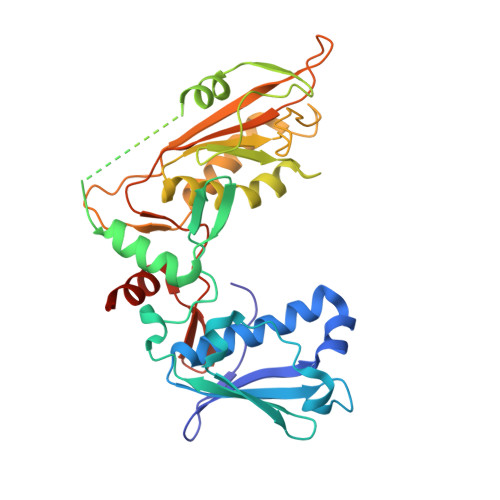
Structural and biochemical characterization of human Schlafen 5.
Metzner, F.J., Huber, E., Hopfner, K.P., Lammens, K.(2022) Nucleic Acids Res
- PubMed: 35037067
- DOI: https://doi.org/10.1093/nar/gkab1278
- Primary Citation of Related Structures:
6RR9, 7PPJ, 7Q3Z - PubMed Abstract:
The Schlafen family belongs to the interferon-stimulated genes and its members are involved in cell cycle regulation, T cell quiescence, inhibition of viral replication, DNA-repair and tRNA processing. Here, we present the cryo-EM structure of full-length human Schlafen 5 (SLFN5) and the high-resolution crystal structure of the highly conserved N-terminal core domain. We show that the core domain does not resemble an ATPase-like fold and neither binds nor hydrolyzes ATP. SLFN5 binds tRNA as well as single- and double-stranded DNA, suggesting a potential role in transcriptional regulation. Unlike rat Slfn13 or human SLFN11, human SLFN5 did not cleave tRNA. Based on the structure, we identified two residues in proximity to the zinc finger motif that decreased DNA binding when mutated. These results indicate that Schlafen proteins have divergent enzymatic functions and provide a structural platform for future biochemical and genetic studies.
- Department of Biochemistry, Gene Center, Feodor-Lynen-Straße 25, 81377 München, Germany.
Organizational Affiliation: