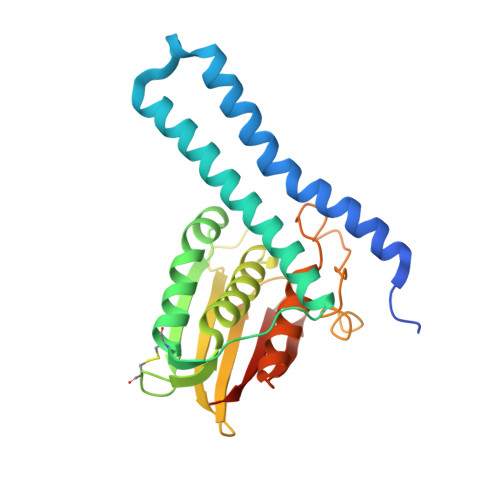
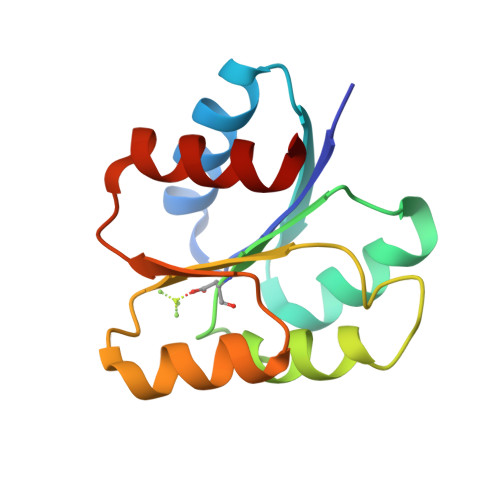
Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Mideros-Mora, C., Miguel-Romero, L., Felipe-Ruiz, A., Casino, P., Marina, A.(2020) Nat Commun 11: 769-769
- PubMed: 32034139
- DOI: https://doi.org/10.1038/s41467-020-14540-5
- Primary Citation of Related Structures:
6RFV, 6RGY, 6RGZ, 6RH0, 6RH1, 6RH2, 6RH7, 6RH8 - PubMed Abstract:
Histidine is a versatile residue playing key roles in enzyme catalysis thanks to the chemistry of its imidazole group that can serve as nucleophile, general acid or base depending on its protonation state. In bacteria, signal transduction relies on two-component systems (TCS) which comprise a sensor histidine kinase (HK) containing a phosphorylatable catalytic His with phosphotransfer and phosphatase activities over an effector response regulator. Recently, a pH-gated model has been postulated to regulate the phosphatase activity of HisKA HKs based on the pH-dependent rotamer switch of the phosphorylatable His. Here, we have revisited this model from a structural and functional perspective on HK853-RR468 and EnvZ-OmpR TCS, the prototypical HisKA HKs. We have found that the rotamer of His is not influenced by the environmental pH, ruling out a pH-gated model and confirming that the chemistry of the His is responsible for the decrease in the phosphatase activity at acidic pH.
- Instituto de Biomedicina de Valencia, Consejo Superior de Investigaciones Científicas (IBV-CSIC), Jaume Roig 11, 46010, Valencia, Spain.
Organizational Affiliation: