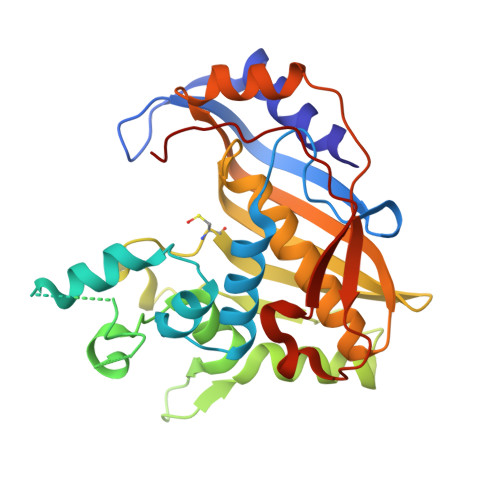
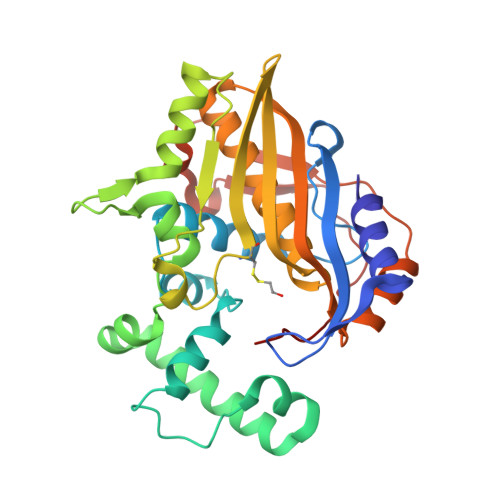
Structural Comparison ofEnterococcus faecalisand Human Thymidylate Synthase Complexes with the Substrate dUMP and Its Analogue FdUMP Provides Hints about Enzyme Conformational Variabilities.
Pozzi, C., Ferrari, S., Luciani, R., Tassone, G., Costi, M.P., Mangani, S.(2019) Molecules 24
- PubMed: 30935102
- DOI: https://doi.org/10.3390/molecules24071257
- Primary Citation of Related Structures:
6QXG, 6QXH, 6QXS, 6QYA - PubMed Abstract:
Thymidylate synthase (TS) is an enzyme of paramount importance as it provides the only de novo source of deoxy-thymidine monophosphate (dTMP). dTMP, essential for DNA synthesis, is produced by the TS-catalyzed reductive methylation of 2'-deoxyuridine-5'-monophosphate (dUMP) using N⁵,N 10 -methylenetetrahydrofolate (mTHF) as a cofactor. TS is ubiquitous and a validated drug target. TS enzymes from different organisms differ in sequence and structure, but are all obligate homodimers. The structural and mechanistic differences between the human and bacterial enzymes are exploitable to obtain selective inhibitors of bacterial TSs that can enrich the currently available therapeutic tools against bacterial infections. Enterococcus faecalis is a pathogen fully dependent on TS for dTMP synthesis. In this study, we present four new crystal structures of Enterococcus faecalis and human TSs in complex with either the substrate dUMP or the inhibitor FdUMP. The results provide new clues about the half-site reactivity of Enterococcus faecalis TS and the mechanisms underlying the conformational changes occurring in the two enzymes. We also identify relevant differences in cofactor and inhibitor binding between Enterococcus faecalis and human TS that can guide the design of selective inhibitors against bacterial TSs.
- Department of Biotechnology, Chemistry and Pharmacy-Department of Excellence 2018-2020, University of Siena, via Aldo Moro 2, 53100 Siena, Italy. pozzi4@unisi.it.
Organizational Affiliation: