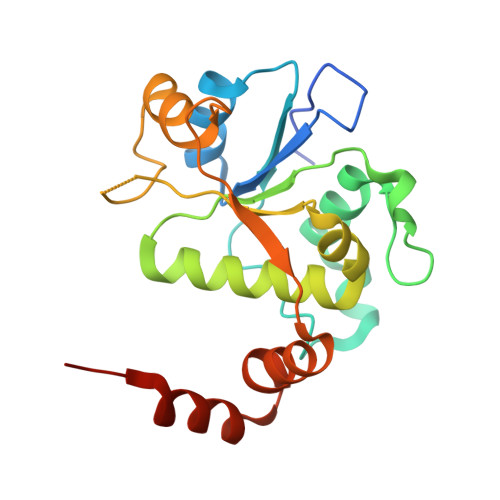
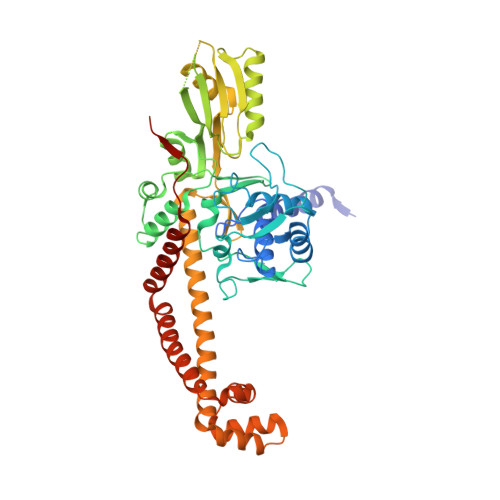
Mechanistic and Structural Basis for the Actions of the Antibacterial Gepotidacin against Staphylococcus aureus Gyrase.
Gibson, E.G., Bax, B., Chan, P.F., Osheroff, N.(2019) ACS Infect Dis 5: 570-581
- PubMed: 30757898
- DOI: https://doi.org/10.1021/acsinfecdis.8b00315
- Primary Citation of Related Structures:
6QTK, 6QTP - PubMed Abstract:
Gepotidacin is a first-in-class triazaacenaphthylene novel bacterial topoisomerase inhibitor (NBTI). The compound has successfully completed phase II trials for the treatment of acute bacterial skin/skin structure infections and for the treatment of uncomplicated urogenital gonorrhea. It also displays robust in vitro activity against a range of wild-type and fluoroquinolone-resistant bacteria. Due to the clinical promise of gepotidacin, a detailed understanding of its interactions with its antibacterial targets is essential. Thus, we characterized the mechanism of action of gepotidacin against Staphylococcus aureus gyrase. Gepotidacin was a potent inhibitor of gyrase-catalyzed DNA supercoiling (IC 50 ≈ 0.047 μM) and relaxation of positively supercoiled substrates (IC 50 ≈ 0.6 μM). Unlike fluoroquinolones, which induce primarily double-stranded DNA breaks, gepotidacin induced high levels of gyrase-mediated single-stranded breaks. No double-stranded breaks were observed even at high gepotidacin concentration, long cleavage times, or in the presence of ATP. Moreover, gepotidacin suppressed the formation of double-stranded breaks. Gepotidacin formed gyrase-DNA cleavage complexes that were stable for >4 h. In vitro competition suggests that gyrase binding by gepotidacin and fluoroquinolones are mutually exclusive. Finally, we determined crystal structures of gepotidacin with the S. aureus gyrase core fusion truncate with nicked (2.31 Å resolution) or intact (uncleaved) DNA (2.37 Å resolution). In both cases, a single gepotidacin molecule was bound midway between the two scissile DNA bonds and in a pocket between the two GyrA subunits. A comparison of the two structures demonstrates conformational flexibility within the central linker of gepotidacin, which may contribute to the activity of the compound.
- Medicines Discovery Institute , Cardiff University , Main Building, Park Place , Cardiff CF10 3AT , United Kingdom.
Organizational Affiliation: