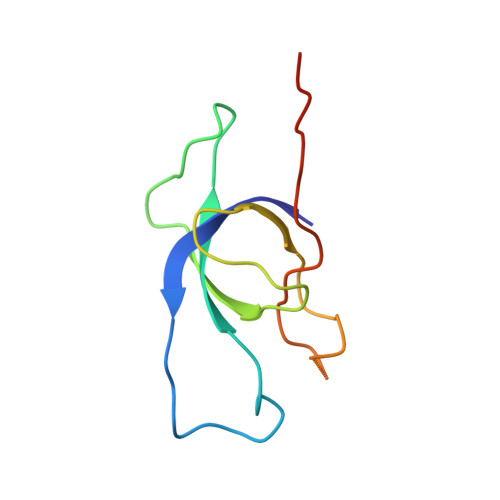
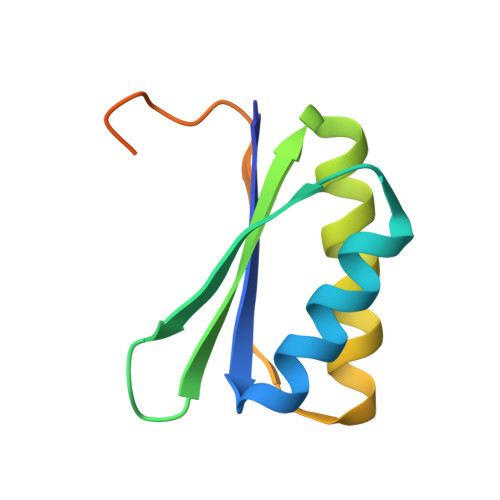
Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles.
Kalnins, G., Cesle, E.E., Jansons, J., Liepins, J., Filimonenko, A., Tars, K.(2020) Nat Commun 11: 388-388
- PubMed: 31959751
- DOI: https://doi.org/10.1038/s41467-019-14205-y
- Primary Citation of Related Structures:
6QN1 - PubMed Abstract:
Bacterial microcompartments (BMCs) are prokaryotic organelles consisting of a protein shell and an encapsulated enzymatic core. BMCs are involved in several biochemical processes, such as choline, glycerol and ethanolamine degradation and carbon fixation. Since non-native enzymes can also be encapsulated in BMCs, an improved understanding of BMC shell assembly and encapsulation processes could be useful for synthetic biology applications. Here we report the isolation and recombinant expression of BMC structural genes from the Klebsiella pneumoniae GRM2 locus, the investigation of mechanisms behind encapsulation of the core enzymes, and the characterization of shell particles by cryo-EM. We conclude that the enzymatic core is encapsulated in a hierarchical manner and that the CutC choline lyase may play a secondary role as an adaptor protein. We also present a cryo-EM structure of a pT = 4 quasi-symmetric icosahedral shell particle at 3.3 Å resolution, and demonstrate variability among the minor shell forms.
- Latvian Biomedical Research and Study Centre, Ratsupites 1, Riga, 1067, Latvia. gints@biomed.lu.lv.
Organizational Affiliation: