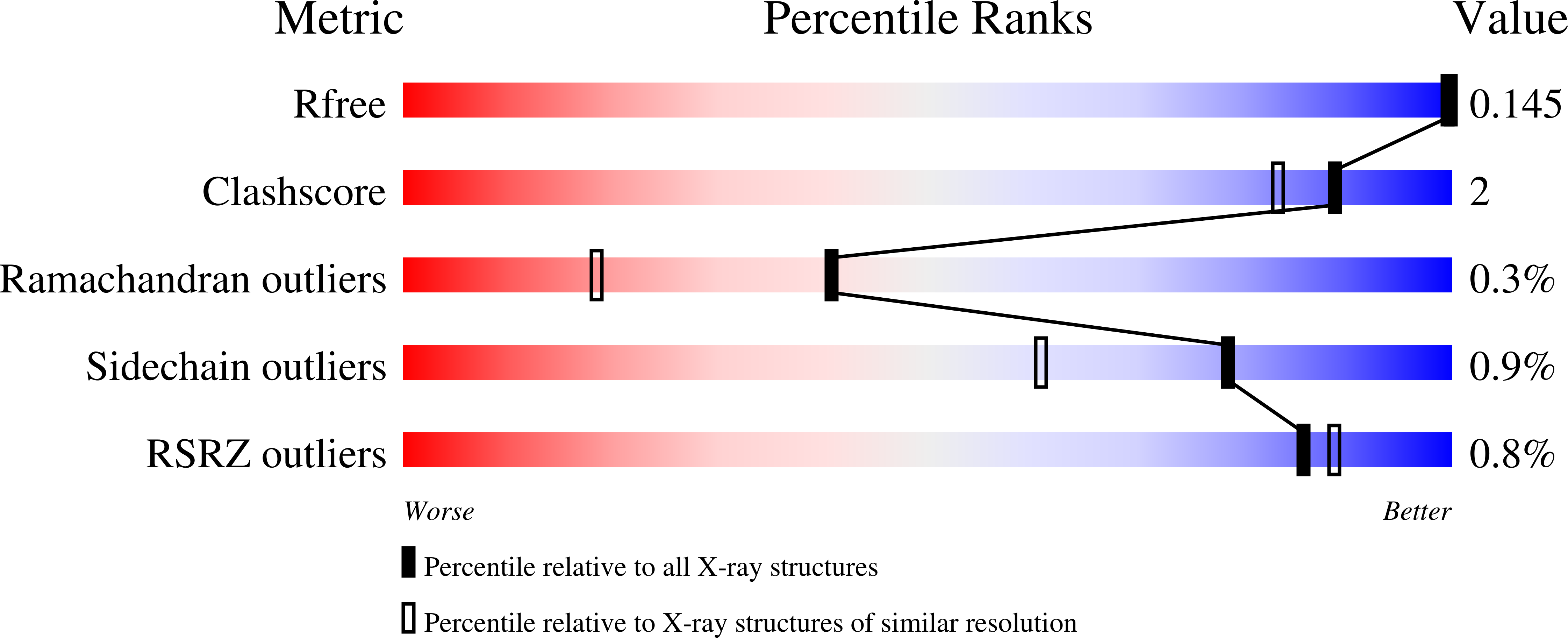
Structural insights into human Arginase-1 pH dependence and its inhibition by the small molecule inhibitor CB-1158.
Grobben, Y., Uitdehaag, J.C.M., Willemsen-Seegers, N., Tabak, W.W.A., de Man, J., Buijsman, R.C., Zaman, G.J.R.(2020) J Struct Biol X 4: 100014-100014
- PubMed: 32647818
- DOI: https://doi.org/10.1016/j.yjsbx.2019.100014
- Primary Citation of Related Structures:
6Q92, 6Q9P, 6QAF - PubMed Abstract:
Arginase-1 is a manganese-dependent metalloenzyme that catalyzes the hydrolysis of L-arginine into L-ornithine and urea. Arginase-1 is abundantly expressed by tumor-infiltrating myeloid cells that promote tumor immunosuppression, which is relieved by inhibition of Arginase-1. We have characterized the potencies of the Arginase-1 reference inhibitors (2 S )-2-amino-6-boronohexanoic acid (ABH) and N ω -hydroxy-nor-L-arginine (nor-NOHA), and studied their pH-dependence and binding kinetics. To gain a better understanding of the structural changes underlying the high pH optimum of Arginase-1 and its pH-dependent inhibition, we determined the crystal structure of the human Arginase-1/ABH complex at pH 7.0 and 9.0. These structures revealed that at increased pH, the manganese cluster assumes a more symmetrical coordination structure, which presumably contributes to its increase in catalytic activity. Furthermore, we show that binding of ABH involves the presence of a sodium ion close to the manganese cluster. We also studied the investigational new drug CB-1158 (INCB001158). This inhibitor has a low-nanomolar potency at pH 7.4 and increases the thermal stability of Arginase-1 more than ABH and nor-NOHA. Moreover, CB-1158 displays slow association and dissociation kinetics at both pH 9.5 and 7.4, as indicated by surface plasmon resonance. The potent character of CB-1158 is presumably due to its increased rigidity compared to ABH as well as the formation of an additional hydrogen-bond network as observed by resolution of the Arginase-1/CB-1158 crystal structure.
Organizational Affiliation:
Netherlands Translational Research Center B.V., Kloosterstraat 9, 5349 AB Oss, The Netherlands.