Tetrahydrofolate Riboswitches Provide Distinct Genetic Outputs to Synthetic and Natural Signals.
Vincent, H.A., Leigh, J., Robinson, C.J., Dunstan, M.S., Ferrer Rios, M.G., Micklefield, J.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| tetrahydrofolate riboswitch aptamer | 89 | Streptococcus mutans UA159 | 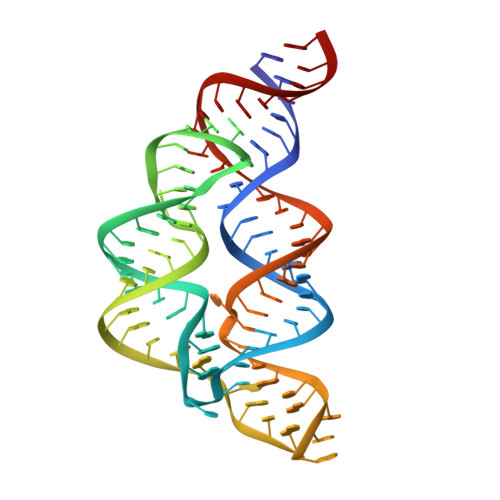 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| T0A Query on T0A | B [auth A], C [auth A] | 5-deazatetrahydropterin C7 H10 N4 O PRQFDDYAWVNJMM-UHFFFAOYSA-N |  | ||
| MG Query on MG | D [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 26.863 | α = 90 |
| b = 68.697 | β = 90 |
| c = 157.309 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| xia2 | data reduction |
| xia2 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| United Kingdom | -- |