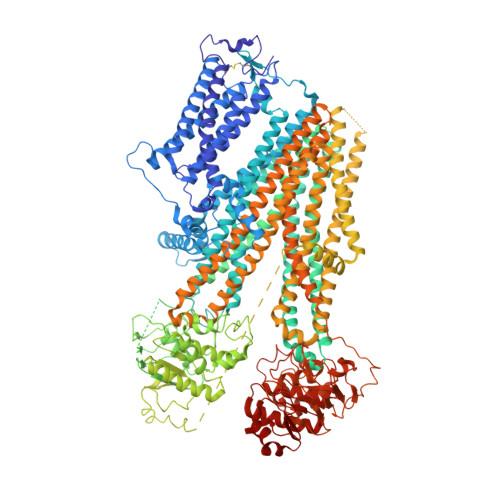
Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Martin, G.M., Sung, M.W., Yang, Z., Innes, L.M., Kandasamy, B., David, L.L., Yoshioka, C., Shyng, S.L.(2019) Elife 8
- PubMed: 31343405
- DOI: https://doi.org/10.7554/eLife.46417
- Primary Citation of Related Structures:
6PZ9, 6PZA, 6PZB, 6PZC, 6PZI - PubMed Abstract:
ATP-sensitive potassium (K ATP ) channels composed of a pore-forming Kir6.2 potassium channel and a regulatory ABC transporter sulfonylurea receptor 1 (SUR1) regulate insulin secretion in pancreatic β-cells to maintain glucose homeostasis. Mutations that impair channel folding or assembly prevent cell surface expression and cause congenital hyperinsulinism. Structurally diverse K ATP inhibitors are known to act as pharmacochaperones to correct mutant channel expression, but the mechanism is unknown. Here, we compare cryoEM structures of a mammalian K ATP channel bound to pharmacochaperones glibenclamide, repaglinide, and carbamazepine. We found all three drugs bind within a common pocket in SUR1. Further, we found the N-terminus of Kir6.2 inserted within the central cavity of the SUR1 ABC core, adjacent the drug binding pocket. The findings reveal a common mechanism by which diverse compounds stabilize the Kir6.2 N-terminus within SUR1's ABC core, allowing it to act as a firm 'handle' for the assembly of metastable mutant SUR1-Kir6.2 complexes.
- Department of Biochemistry and Molecular Biology, Oregon Health & Science University, Portland, United States.
Organizational Affiliation: