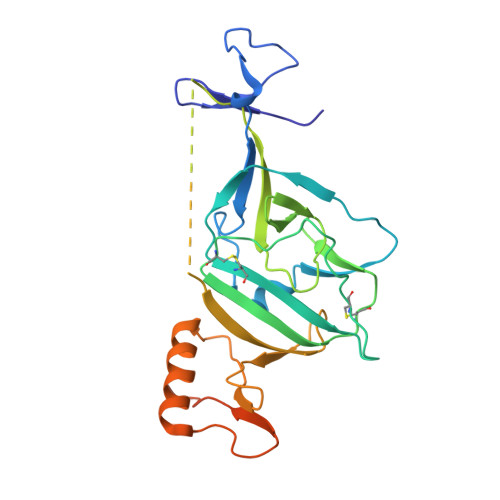
Analysis of a Therapeutic Antibody Cocktail Reveals Determinants for Cooperative and Broad Ebolavirus Neutralization.
Gilchuk, P., Murin, C.D., Milligan, J.C., Cross, R.W., Mire, C.E., Ilinykh, P.A., Huang, K., Kuzmina, N., Altman, P.X., Hui, S., Gunn, B.M., Bryan, A.L., Davidson, E., Doranz, B.J., Turner, H.L., Alkutkar, T., Flinko, R., Orlandi, C., Carnahan, R., Nargi, R., Bombardi, R.G., Vodzak, M.E., Li, S., Okoli, A., Ibeawuchi, M., Ohiaeri, B., Lewis, G.K., Alter, G., Bukreyev, A., Saphire, E.O., Geisbert, T.W., Ward, A.B., Crowe Jr., J.E.(2020) Immunity 52: 388-403.e12
- PubMed: 32023489
- DOI: https://doi.org/10.1016/j.immuni.2020.01.001
- Primary Citation of Related Structures:
6OZ9, 6PCI, 6UYE - PubMed Abstract:
Structural principles underlying the composition of protective antiviral monoclonal antibody (mAb) cocktails are poorly defined. Here, we exploited antibody cooperativity to develop a therapeutic mAb cocktail against Ebola virus. We systematically analyzed the antibody repertoire in human survivors and identified a pair of potently neutralizing mAbs that cooperatively bound to the ebolavirus glycoprotein (GP). High-resolution structures revealed that in a two-antibody cocktail, molecular mimicry was a major feature of mAb-GP interactions. Broadly neutralizing mAb rEBOV-520 targeted a conserved epitope on the GP base region. mAb rEBOV-548 bound to a glycan cap epitope, possessed neutralizing and Fc-mediated effector function activities, and potentiated neutralization by rEBOV-520. Remodeling of the glycan cap structures by the cocktail enabled enhanced GP binding and virus neutralization. The cocktail demonstrated resistance to virus escape and protected non-human primates (NHPs) against Ebola virus disease. These data illuminate structural principles of antibody cooperativity with implications for development of antiviral immunotherapeutics.
- Vanderbilt Vaccine Center, Vanderbilt University Medical Center, Nashville, TN 37232, USA.
Organizational Affiliation: