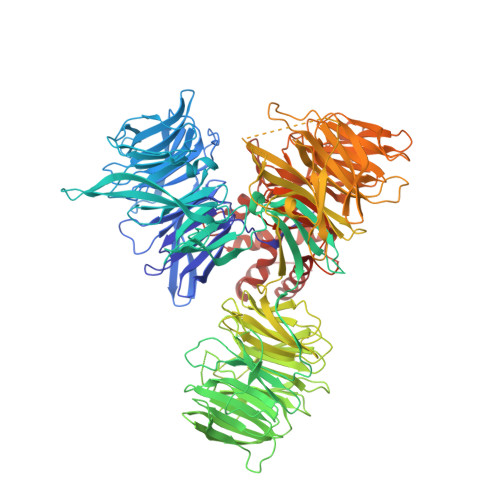
Structural Basis and Kinetic Pathway of RBM39 Recruitment to DCAF15 by a Sulfonamide Molecular Glue E7820.
Du, X., Volkov, O.A., Czerwinski, R.M., Tan, H., Huerta, C., Morton, E.R., Rizzi, J.P., Wehn, P.M., Xu, R., Nijhawan, D., Wallace, E.M.(2019) Structure 27: 1625-1633.e3
- PubMed: 31693911
- DOI: https://doi.org/10.1016/j.str.2019.10.005
- Primary Citation of Related Structures:
6PAI - PubMed Abstract:
E7820 and indisulam are two examples of aryl sulfonamides that recruit RBM39 to Rbx-Cul4-DDA1-DDB1-DCAF15 E3 ligase complex, leading to its ubiquitination and degradation by the proteasome. To understand their mechanism of action, we performed kinetic analysis on the recruitment of RBM39 to DCAF15 and solved a crystal structure of DDA1-DDB1-DCAF15 in complex with E7820 and the RRM2 domain of RBM39. E7820 packs in a shallow pocket on the surface of DCAF15 and the resulting modified interface binds RBM39 through the α1 helix of the RRM2 domain. Our kinetic studies revealed that aryl sulfonamide and RBM39 bind to DCAF15 in a synergistic manner. The structural and kinetic studies confirm aryl sulfonamides as molecular glues in the recruitment of RBM39 and provide a framework for future efforts to utilize DCAF15 to degrade other proteins of interest.
- Peloton Therapeutics, Dallas, Texas, a Subsidiary of Merck & Co., Inc., Kenilworth, NJ 07033, USA. Electronic address: xinlin.du@merck.com.
Organizational Affiliation: