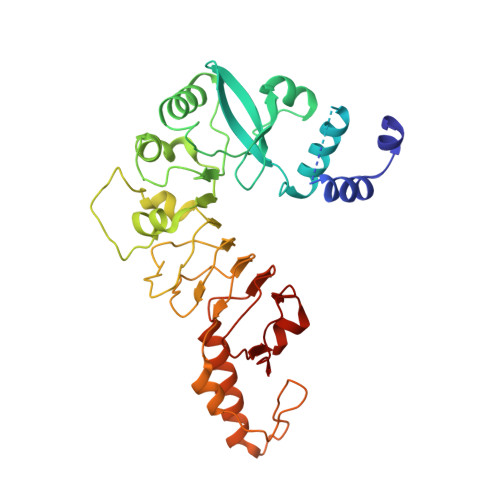
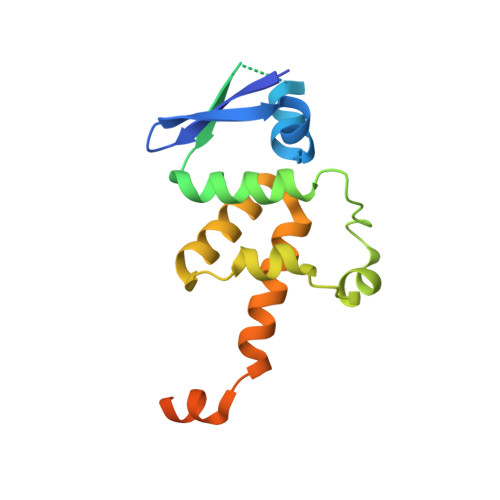
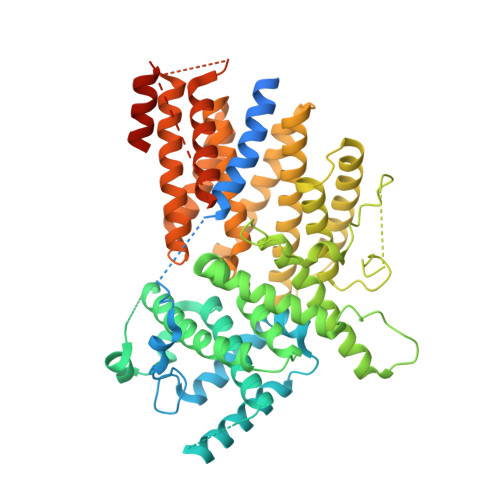
Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
Lee, P.D., Wei, H., Tan, D., Harrison, S.C.(2019) J Mol Biology 431: 4444-4454
- PubMed: 31425683
- DOI: https://doi.org/10.1016/j.jmb.2019.08.003
- Primary Citation of Related Structures:
6P7V, 6P7W, 6P7X - PubMed Abstract:
Kinetochores are the multiprotein complexes that link chromosomal centromeres to mitotic-spindle microtubules. Budding yeast centromeres comprise three sequential "centromere-determining elements", CDEI, II, and III. CDEI (8 bp) and CDEIII (∼25 bp) are conserved between Kluyveromyces lactis and Saccharomyces cerevisiae, but CDEII in the former is twice as long (160 bp) as CDEII in the latter (80 bp). The CBF3 complex recognizes CDEIII and is required for assembly of a centromeric nucleosome, which in turn recruits other kinetochore components. To understand differences in centromeric nucleosome assembly between K. lactis and S. cerevisiae, we determined the structure of a K. lactis CBF3 complex by electron cryomicroscopy at ∼4 Å resolution and compared it with published structures of S. cerevisiae CBF3. We show differences in the pose of Ndc10 and discuss potential models of the K. lactis centromeric nucleosome that account for the extended CDEII length.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA, 02115, USA; Graduate Program in Virology, Harvard Medical School, Boston, MA, 02115, USA.
Organizational Affiliation: