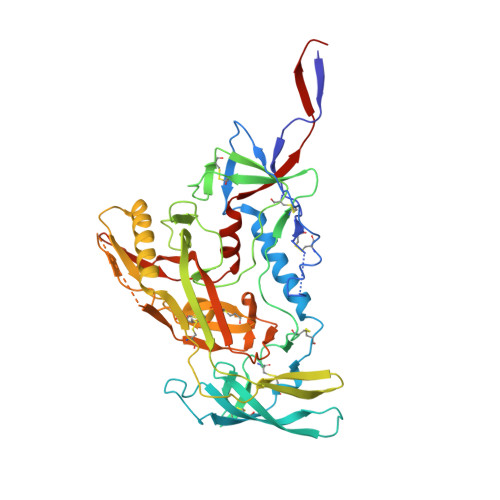
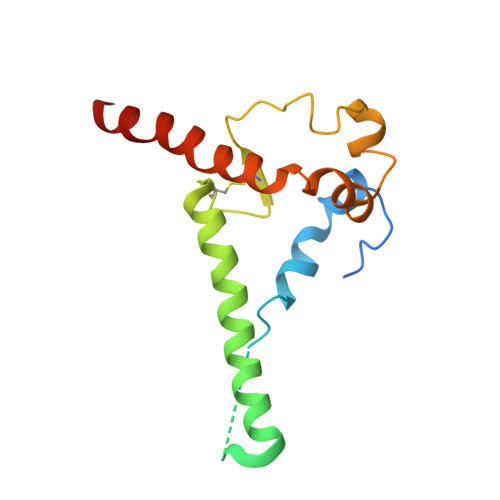
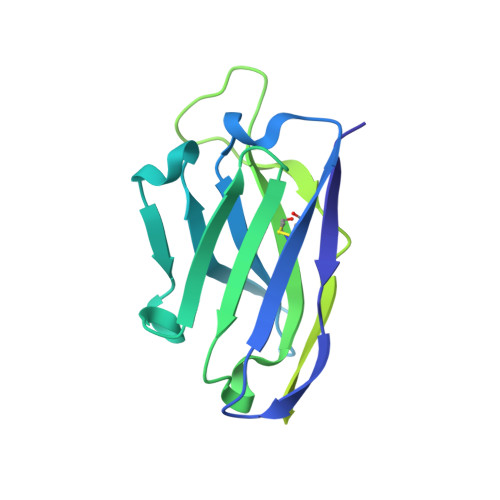
Networks of HIV-1 Envelope Glycans Maintain Antibody Epitopes in the Face of Glycan Additions and Deletions.
Seabright, G.E., Cottrell, C.A., van Gils, M.J., D'addabbo, A., Harvey, D.J., Behrens, A.J., Allen, J.D., Watanabe, Y., Scaringi, N., Polveroni, T.M., Maker, A., Vasiljevic, S., de Val, N., Sanders, R.W., Ward, A.B., Crispin, M.(2020) Structure 28: 897
- PubMed: 32433992
- DOI: https://doi.org/10.1016/j.str.2020.04.022
- Primary Citation of Related Structures:
6OZC - PubMed Abstract:
Numerous broadly neutralizing antibodies (bnAbs) have been identified that target the glycans of the HIV-1 envelope spike. Neutralization breadth is notable given that glycan processing can be substantially influenced by the presence or absence of neighboring glycans. Here, using a stabilized recombinant envelope trimer, we investigate the degree to which mutations in the glycan network surrounding an epitope impact the fine glycan processing of antibody targets. Using cryo-electron microscopy and site-specific glycan analysis, we reveal the importance of glycans in the formation of the 2G12 bnAb epitope and show that the epitope is only subtly impacted by variations in the glycan network. In contrast, we show that the PG9 and PG16 glycan-based epitopes at the trimer apex are dependent on the presence of the highly conserved surrounding glycans. Glycan networks underpin the conservation of bnAb epitopes and are an important parameter in immunogen design.
- School of Biological Sciences, University of Southampton, Southampton SO17 1BJ, UK; Oxford Glycobiology Institute, Department of Biochemistry, University of Oxford, Oxford OX1 3QU, UK.
Organizational Affiliation: