A structural mechanism for phosphorylation-dependent inactivation of the AP2 complex.
Partlow, E.A., Baker, R.W., Beacham, G.M., Chappie, J.S., Leschziner, A.E., Hollopeter, G.(2019) Elife 8
- PubMed: 31464684
- DOI: https://doi.org/10.7554/eLife.50003
- Primary Citation of Related Structures:
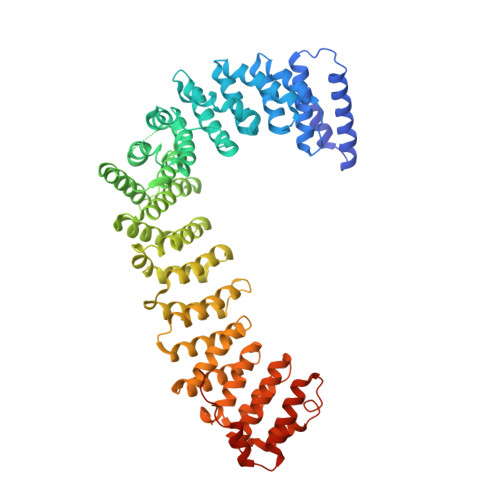
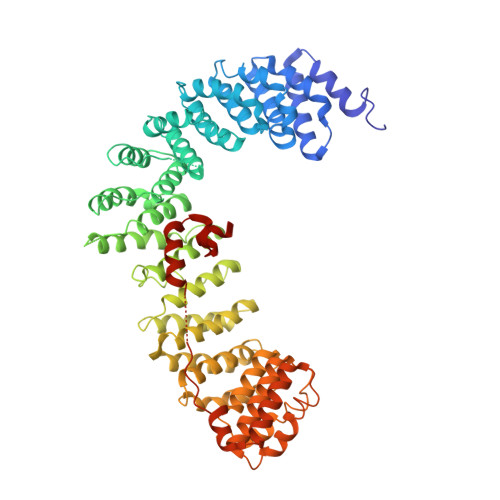
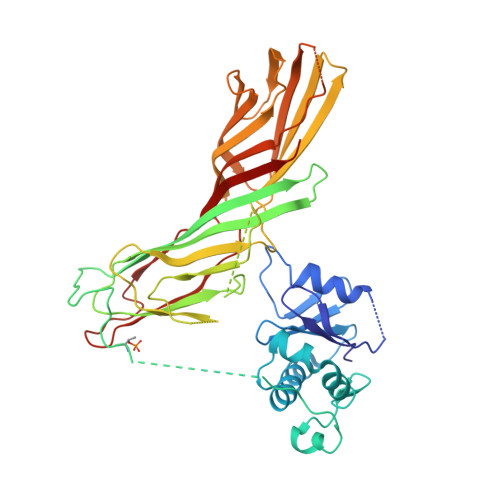
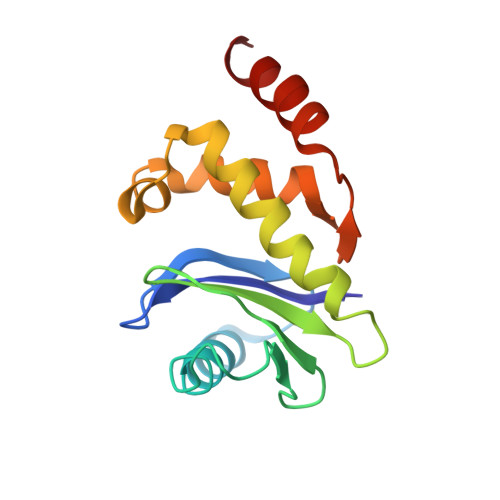
6OWO, 6OXL - PubMed Abstract:
Endocytosis of transmembrane proteins is orchestrated by the AP2 clathrin adaptor complex. AP2 dwells in a closed, inactive state in the cytosol, but adopts an open, active conformation on the plasma membrane. Membrane-activated complexes are also phosphorylated, but the significance of this mark is debated. We recently proposed that NECAP negatively regulates AP2 by binding open and phosphorylated complexes (Beacham et al., 2018). Here, we report high-resolution cryo-EM structures of NECAP bound to phosphorylated AP2. The site of AP2 phosphorylation is directly coordinated by residues of the NECAP PHear domain that are predicted from genetic screens in C. elegans . Using membrane mimetics to generate conformationally open AP2, we find that a second domain of NECAP binds these complexes and cryo-EM reveals both domains of NECAP engaging closed, inactive AP2. Assays in vitro and in vivo confirm these domains cooperate to inactivate AP2. We propose that phosphorylation marks adaptors for inactivation.
- Department of Molecular Medicine, Cornell University, New York, United States.
Organizational Affiliation: