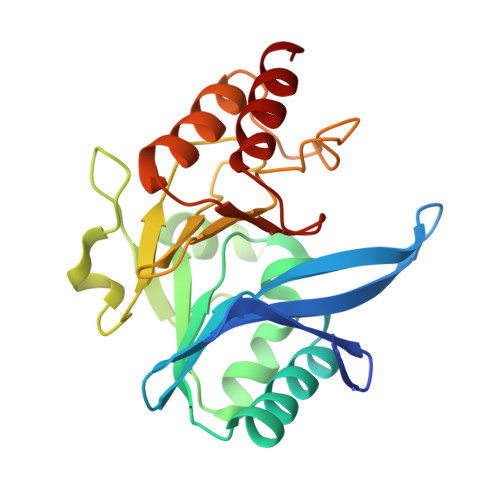
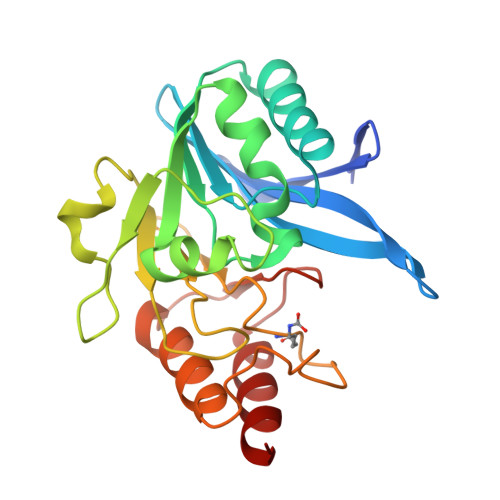
A Lysine-Targeted Affinity Label for Serine-beta-Lactamase Also Covalently Modifies New Delhi Metallo-beta-lactamase-1 (NDM-1).
Thomas, P.W., Cammarata, M., Brodbelt, J.S., Monzingo, A.F., Pratt, R.F., Fast, W.(2019) Biochemistry 58: 2834-2843
- PubMed: 31145588
- DOI: https://doi.org/10.1021/acs.biochem.9b00393
- Primary Citation of Related Structures:
6OVZ - PubMed Abstract:
The divergent sequences, protein structures, and catalytic mechanisms of serine- and metallo-β-lactamases hamper the development of wide-spectrum β-lactamase inhibitors that can block both types of enzymes. The O-aryloxycarbonyl hydroxamate inactivators of Enterobacter cloacae P99 class C serine-β-lactamase are unusual covalent inhibitors in that they target both active-site Ser and Lys residues, resulting in a cross-link consisting of only two atoms. Many clinically relevant metallo-β-lactamases have an analogous active-site Lys residue used to bind β-lactam substrates, suggesting a common site to target with covalent inhibitors. Here, we demonstrate that an O-aryloxycarbonyl hydroxamate inactivator of serine-β-lactamases can also serve as a classical affinity label for New Delhi metallo-β-lactamase-1 (NDM-1). Rapid dilution assays, site-directed mutagenesis, and global kinetic fitting are used to map covalent modification at Lys211 and determine K I (140 μM) and k inact (0.045 min -1 ) values. Mass spectrometry of the intact protein and the use of ultraviolet photodissociation for extensive fragmentation confirm stoichiometric covalent labeling that occurs specifically at Lys211. A 2.0 Å resolution X-ray crystal structure of inactivated NDM-1 reveals that the covalent adduct is bound at the substrate-binding site but is not directly coordinated to the active-site zinc cluster. These results indicate that Lys-targeted affinity labels might be a successful strategy for developing compounds that can inactivate both serine- and metallo-β-lactamases.
- Department of Chemistry , Wesleyan University , Middletown , Connecticut 06459 , United States.
Organizational Affiliation: