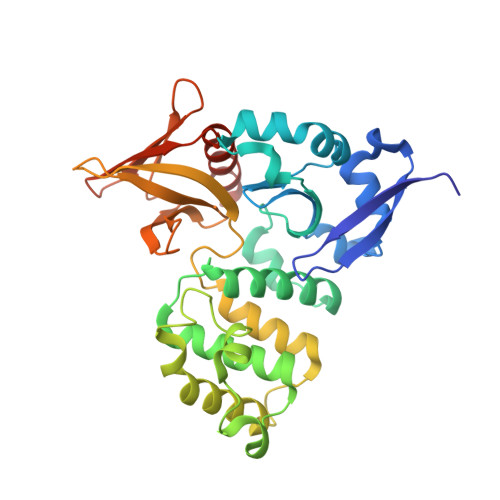
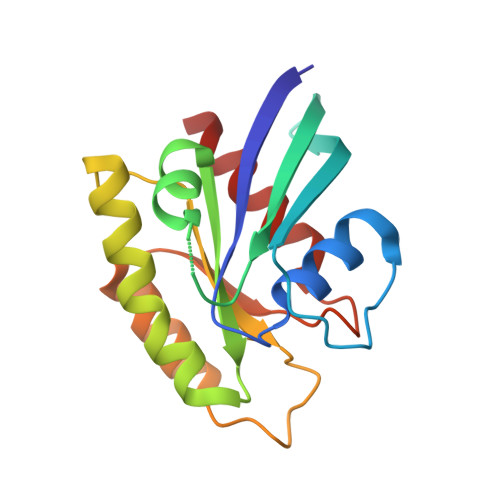
Inhibition of the HEG1-KRIT1 interaction increases KLF4 and KLF2 expression in endothelial cells
Lopez-Ramirez, M.A., McCurdy, S., Li, W., Haynes, M.K., Hale, P., Francisco, K., Oukoloff, K., Bautista, M., Choi, C.H., Sun, H., Gongol, B., Shyy, J.Y., Ballatore, C., Sklar, L.A., Gingras, A.R.(2021) FASEB Bioadv