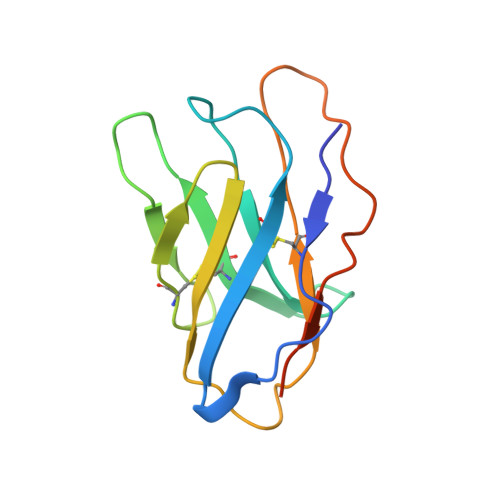
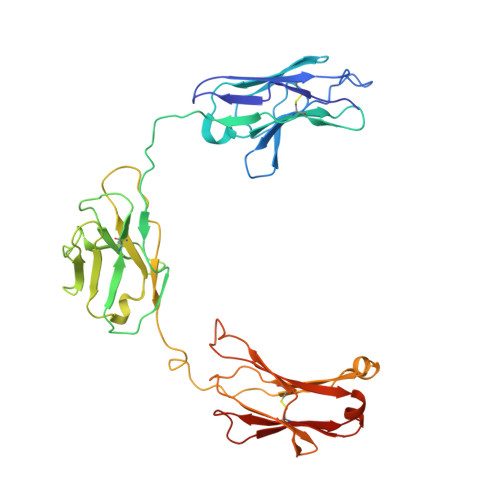
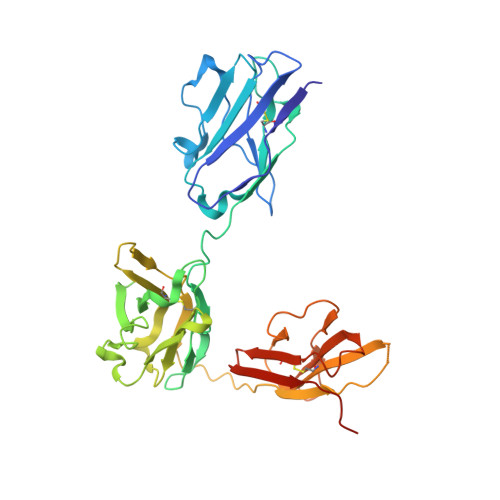
Trispecific antibodies enhance the therapeutic efficacy of tumor-directed T cells through T cell receptor co-stimulation
Wu, L., Seung, E., Xu, L., Rao, E., Lord, D.M., Wei, R.R., Cortez-Retamozo, V., Ospina, B., Posternak, V., Ulinski, G., Piepenhagen, P., Francesconi, E., El Murr, N., Beil, C., Dabdoubi, T., Cameron, B., Bertrand, T., Ferrari, P., Pouzieux, S., Lemoine, C., Prades, C., Park, A., Qiu, H., Song, Z., Zhang, B., Sun, F., Chiron, M., Rao, S., Radosevic, K., Zhi-yong, Y., Nabel, G.J.To be published.