A Dissection of Oligomerization by the TRIM28 Tripartite Motif and the Interaction with Members of the Krab-ZFP Family.
Sun, Y., Keown, J.R., Black, M.M., Raclot, C., Demarais, N., Trono, D., Turelli, P., Goldstone, D.C.(2019) J Mol Biol 431: 2511-2527
- PubMed: 31078555
- DOI: https://doi.org/10.1016/j.jmb.2019.05.002
- Primary Citation of Related Structures:
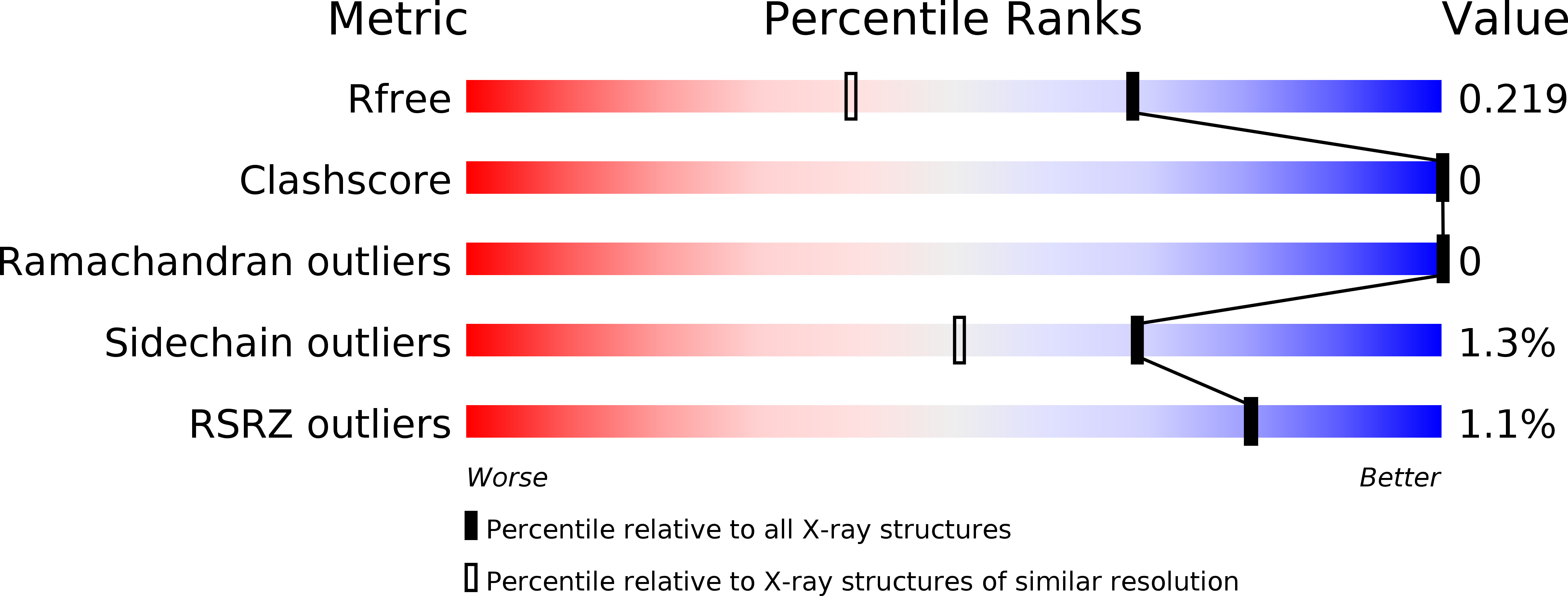
6O5K - PubMed Abstract:
TRIM28 (also known as KAP1 or TIF1β) is the universal co-repressor of the Krüppel-associated box-containing zinc finger proteins (Krab-ZFPs), the largest family of transcription factors in mammals. During early embryogenesis, TRIM28 mediates the transcriptional silencing of many endogenous retroviral elements and genomic imprinted sites. Silencing is initiated by the recruitment of TRIM28 to a target locus by members of the Krab-ZFP. Subsequently, TRIM28 functions as a scaffold protein to recruit chromatin modifying effectors featuring SETDB1, HP1 and the NuRD complex. Although many protein partners involved in silencing have been identified, the molecular basis of the protein interactions that mediate silencing remains largely unclear. In the present study, we identified the first Bbox domain (T28_B1 135-203) as a molecular interface responsible for the formation of higher-order oligomers of TRIM28. The structure of this domain reveals a new interface on the surface of the Bbox domain. Mutants disrupting the interface disrupt the formation of oligomers but have no observed effect on transcriptional silencing defining a single TRIM28 dimer as the functional unit for silencing. Using assembly-deficient mutants, we employed small-angle X-ray scattering and biophysical techniques to characterize binding to member of the Krab-ZFP family. This allows us to narrow and define the binding interface to the center of the coiled-coil region (residues 294-321) of TRIM28 and define mutants that abolish binding to the Krab-ZFP proteins.
Organizational Affiliation:
School of Biological Sciences, University of Auckland, Auckland, New Zealand.