Cryo-EM reveals the structural basis of long-range electron transport in a cytochrome-based bacterial nanowire.
Filman, D.J., Marino, S.F., Ward, J.E., Yang, L., Mester, Z., Bullitt, E., Lovley, D.R., Strauss, M.(2019) Commun Biol 2: 219-219
- PubMed: 31240257
- DOI: https://doi.org/10.1038/s42003-019-0448-9
- Primary Citation of Related Structures:
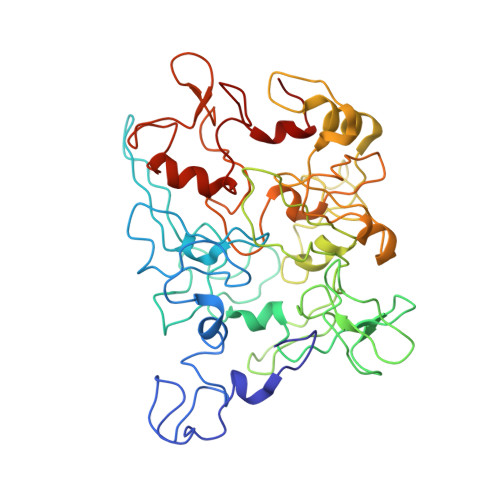
6NEF - PubMed Abstract:
Electrically conductive pili from Geobacter species, termed bacterial nanowires, are intensely studied for their biological significance and potential in the development of new materials. Using cryo-electron microscopy, we have characterized nanowires from conductive G. sulfurreducens pili preparations that are composed solely of head-to-tail stacked monomers of the six-heme C-type cytochrome OmcS. The unique fold of OmcS - closely wrapped around a continuous stack of hemes that can serve as an uninterrupted path for electron transport - generates a scaffold that supports the unbranched chain of hemes along the central axis of the filament. We present here, at 3.4 Å resolution, the structure of this cytochrome-based filament and discuss its possible role in long-range biological electron transport.
- 1Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115 USA.
Organizational Affiliation: