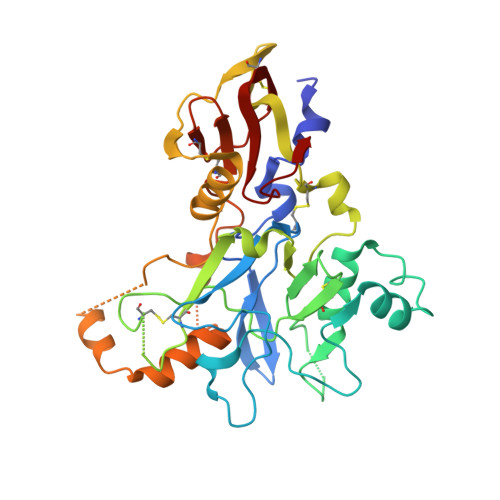
Identification of the Binding Site of Apical Membrane Antigen 1 (AMA1) Inhibitors Using a Paramagnetic Probe.
Akter, M., Drinkwater, N., Devine, S.M., Drew, S.C., Krishnarjuna, B., Debono, C.O., Wang, G., Scanlon, M.J., Scammells, P.J., McGowan, S., MacRaild, C.A., Norton, R.S.(2019) ChemMedChem 14: 603-612
- PubMed: 30653832
- DOI: https://doi.org/10.1002/cmdc.201800802
- Primary Citation of Related Structures:
6N7Q, 6N87 - PubMed Abstract:
Apical membrane antigen 1 (AMA1) is essential for the invasion of host cells by malaria parasites. Several small-molecule ligands have been shown to bind to a conserved hydrophobic cleft in Plasmodium falciparum AMA1. However, a lack of detailed structural information on the binding pose of these molecules has hindered their further optimisation as inhibitors. We have developed a spin-labelled peptide based on RON2, the native binding partner of AMA1, to probe the binding sites of compounds on PfAMA1. The crystal structure of this peptide bound to PfAMA1 shows that it binds at one end of the hydrophobic groove, leaving much of the binding site unoccupied and allowing fragment hits to bind without interference. In paramagnetic relaxation enhancement (PRE)-based NMR screening, the 1 H relaxation rates of compounds binding close to the probe were enhanced. Compounds experienced different degrees of PRE as a result of their different orientations relative to the spin label while bound to AMA1. Thus, PRE-derived distance constraints can be used to identify binding sites and guide further hit optimisation.
- Medicinal Chemistry, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, Victoria, 3052, Australia.
Organizational Affiliation: