Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
Shoffner, G.M., Peng, Z., Guo, F.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA (123-MER) | 123 | Homo sapiens | 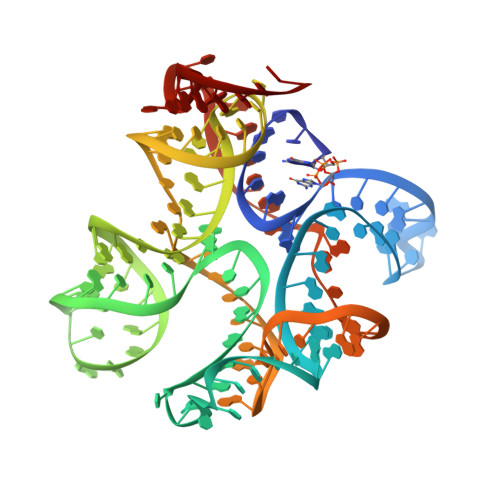 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 2BA Query on 2BA | C [auth A], D [auth A] | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8
]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide C20 H24 N10 O12 P2 PDXMFTWFFKBFIN-XPWFQUROSA-N |  | ||
| 5GP Query on 5GP | B [auth A] | GUANOSINE-5'-MONOPHOSPHATE C10 H14 N5 O8 P RQFCJASXJCIDSX-UUOKFMHZSA-N |  | ||
| SO4 Query on SO4 | J [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| K Query on K | G [auth A] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| MG Query on MG | E [auth A], F [auth A], H [auth A], I [auth A], K [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 114.56 | α = 90 |
| b = 114.56 | β = 90 |
| c = 115.06 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| XDS | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Science Foundation (NSF, United States) | United States | 1616265 |