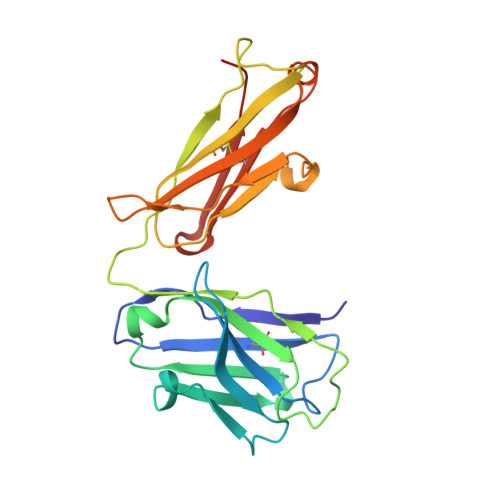
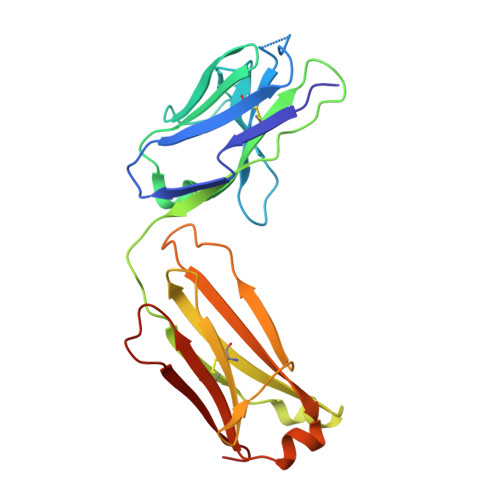
VISTA is an acidic pH-selective ligand for PSGL-1.
Johnston, R.J., Su, L.J., Pinckney, J., Critton, D., Boyer, E., Krishnakumar, A., Corbett, M., Rankin, A.L., Dibella, R., Campbell, L., Martin, G.H., Lemar, H., Cayton, T., Huang, R.Y., Deng, X., Nayeem, A., Chen, H., Ergel, B., Rizzo, J.M., Yamniuk, A.P., Dutta, S., Ngo, J., Shorts, A.O., Ramakrishnan, R., Kozhich, A., Holloway, J., Fang, H., Wang, Y.K., Yang, Z., Thiam, K., Rakestraw, G., Rajpal, A., Sheppard, P., Quigley, M., Bahjat, K.S., Korman, A.J.(2019) Nature 574: 565-570
- PubMed: 31645726
- DOI: https://doi.org/10.1038/s41586-019-1674-5
- Primary Citation of Related Structures:
6MVL - PubMed Abstract:
Co-inhibitory immune receptors can contribute to T cell dysfunction in patients with cancer 1,2 . Blocking antibodies against cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) and programmed cell death 1 (PD-1) partially reverse this effect and are becoming standard of care in an increasing number of malignancies 3 . However, many of the other axes by which tumours become inhospitable to T cells are not fully understood. Here we report that V-domain immunoglobulin suppressor of T cell activation (VISTA) engages and suppresses T cells selectively at acidic pH such as that found in tumour microenvironments. Multiple histidine residues along the rim of the VISTA extracellular domain mediate binding to the adhesion and co-inhibitory receptor P-selectin glycoprotein ligand-1 (PSGL-1). Antibodies engineered to selectively bind and block this interaction in acidic environments were sufficient to reverse VISTA-mediated immune suppression in vivo. These findings identify a mechanism by which VISTA may engender resistance to anti-tumour immune responses, as well as an unexpectedly determinative role for pH in immune co-receptor engagement.
- Immuno-Oncology Discovery, Bristol-Myers Squibb, Redwood City, CA, USA. robert.johnston@bms.com.
Organizational Affiliation: