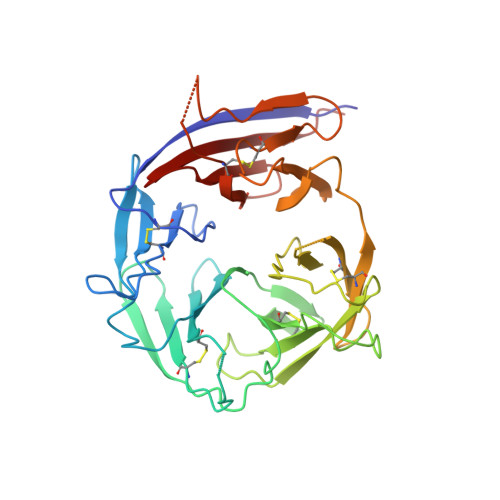
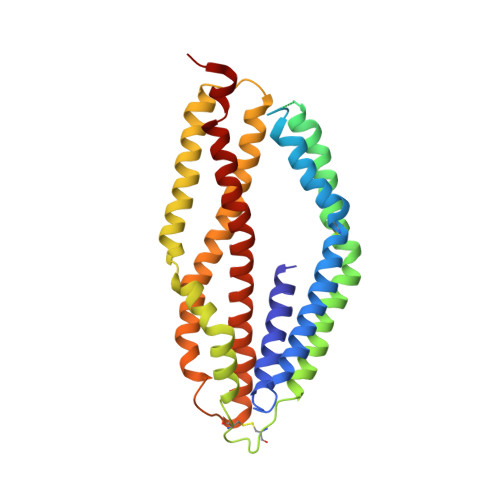
Structure of Plasmodium falciparum Rh5-CyRPA-Ripr invasion complex.
Wong, W., Huang, R., Menant, S., Hong, C., Sandow, J.J., Birkinshaw, R.W., Healer, J., Hodder, A.N., Kanjee, U., Tonkin, C.J., Heckmann, D., Soroka, V., Sogaard, T.M.M., Jorgensen, T., Duraisingh, M.T., Czabotar, P.E., de Jongh, W.A., Tham, W.H., Webb, A.I., Yu, Z., Cowman, A.F.(2019) Nature 565: 118-121
- PubMed: 30542156
- DOI: https://doi.org/10.1038/s41586-018-0779-6
- Primary Citation of Related Structures:
6MPV - PubMed Abstract:
Plasmodium falciparum causes the severe form of malaria that has high levels of mortality in humans. Blood-stage merozoites of P. falciparum invade erythrocytes, and this requires interactions between multiple ligands from the parasite and receptors in hosts. These interactions include the binding of the Rh5-CyRPA-Ripr complex with the erythrocyte receptor basigin 1,2 , which is an essential step for entry into human erythrocytes. Here we show that the Rh5-CyRPA-Ripr complex binds the erythrocyte cell line JK-1 significantly better than does Rh5 alone, and that this binding occurs through the insertion of Rh5 and Ripr into host membranes as a complex with high molecular weight. We report a cryo-electron microscopy structure of the Rh5-CyRPA-Ripr complex at subnanometre resolution, which reveals the organization of this essential invasion complex and the mode of interactions between members of the complex, and shows that CyRPA is a critical mediator of complex assembly. Our structure identifies blades 4-6 of the β-propeller of CyRPA as contact sites for Rh5 and Ripr. The limited contacts between Rh5-CyRPA and CyRPA-Ripr are consistent with the dissociation of Rh5 and Ripr from CyRPA for membrane insertion. A comparision of the crystal structure of Rh5-basigin with the cryo-electron microscopy structure of Rh5-CyRPA-Ripr suggests that Rh5 and Ripr are positioned parallel to the erythrocyte membrane before membrane insertion. This provides information on the function of this complex, and thereby provides insights into invasion by P. falciparum.
- Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria, Australia.
Organizational Affiliation: