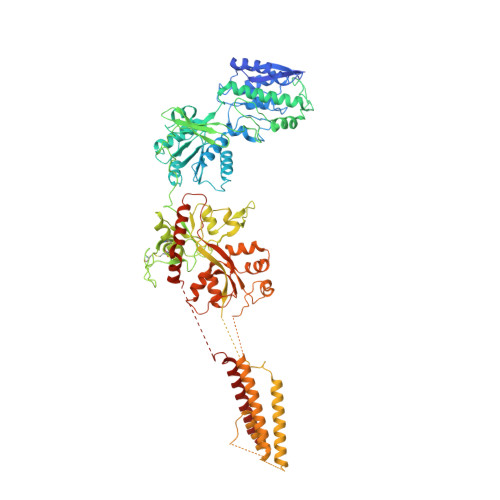
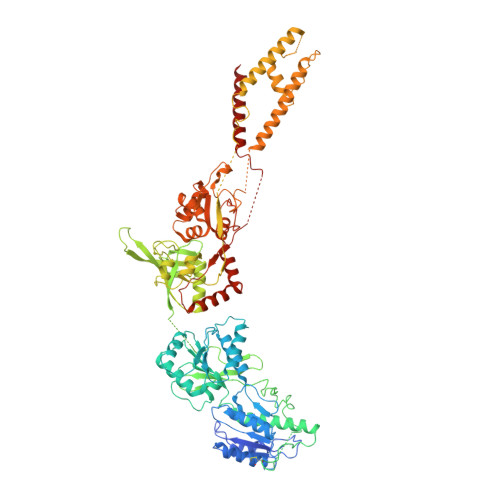
Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Jalali-Yazdi, F., Chowdhury, S., Yoshioka, C., Gouaux, E.(2018) Cell 175: 1520-1532.e15
- PubMed: 30500536
- DOI: https://doi.org/10.1016/j.cell.2018.10.043
- Primary Citation of Related Structures:
6MM9, 6MMA, 6MMB, 6MMG, 6MMH, 6MMI, 6MMJ, 6MMK, 6MML, 6MMM, 6MMN, 6MMP, 6MMR, 6MMS, 6MMT, 6MMU, 6MMV, 6MMW, 6MMX - PubMed Abstract:
N-methyl-D-aspartate receptors (NMDARs) play essential roles in memory formation, neuronal plasticity, and brain development, with their dysfunction linked to a range of disorders from ischemia to schizophrenia. Zinc and pH are physiological allosteric modulators of NMDARs, with GluN2A-containing receptors inhibited by nanomolar concentrations of divalent zinc and by excursions to low pH. Despite the widespread importance of zinc and proton modulation of NMDARs, the molecular mechanism by which these ions modulate receptor activity has proven elusive. Here, we use cryoelectron microscopy to elucidate the structure of the GluN1/GluN2A NMDAR in a large ensemble of conformations under a range of physiologically relevant zinc and proton concentrations. We show how zinc binding to the amino terminal domain elicits structural changes that are transduced though the ligand-binding domain and result in constriction of the ion channel gate.
- Vollum Institute, Oregon Health and Science University, Portland, OR 97239, USA.
Organizational Affiliation: