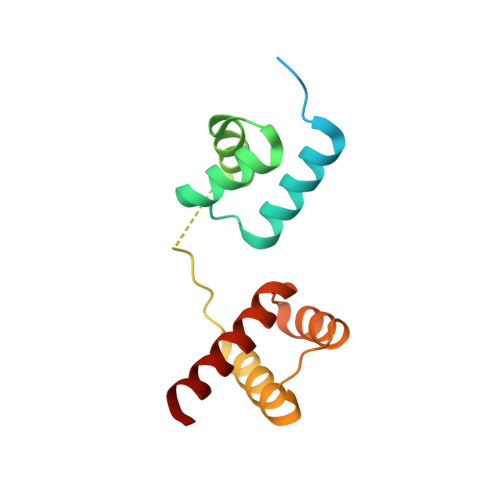
Structural basis for designing an array of engrailed homeodomains.
Sunami, T., Hirano, Y., Tamada, T., Kono, H.(2020) Acta Crystallogr D Struct Biol 76: 824-833
- PubMed: 32876058
- DOI: https://doi.org/10.1107/S2059798320009237
- Primary Citation of Related Structures:
6M3D - PubMed Abstract:
Small DNA-binding proteins that target desired sequences have the potential to act as a scaffold for molecular tools such as genome editing. In this study, an engrailed homeodomain (EHD) was chosen and it was evaluated whether it could be used as a molecular module that can connect to itself to recognize a longer target sequence. It was previously shown that two EHDs connected by a linker (EHD 2 ) recognize a target sequence twice as long as that recognized by a single EHD in cells only when Arg53 in each EHD in the tandem protein is mutated to alanine {(EHD[R53A]) 2 }. To investigate the recognition mechanism of (EHD[R53A]) 2 , the crystal structure of the (EHD[R53A]) 2 -DNA complex was determined at 1.6 Å resolution. The individual EHDs were found to adopt the typical homeodomain fold. Most importantly, the base-specific interactions in the major groove necessary for the affinity/specificity of wild-type EHD were preserved in (EHD[R53A]) 2 . Bacterial assays confirmed that the base-specific interactions are retained under cellular conditions. These observations indicate that the R53A mutation only causes a loss of the arginine-phosphate interaction at the protein-DNA interface, which reduces the DNA-binding affinity compared with the wild type. It is therefore concluded that (EHD[R53A]) 2 precisely recognizes tandem target sites within cells, enabling the individual EHDs to concurrently bind to the target sites with modest binding affinity. This suggests that modulation of the binding activity of each EHD is vital to construct a protein array that can precisely recognize a sequence with multiple target sites.
- Molecular Modeling and Simulation Group, National Institutes for Quantum and Radiological Science and Technology, 8-1-7 Umemidai, Kizugawa 619-0215, Japans.
Organizational Affiliation: