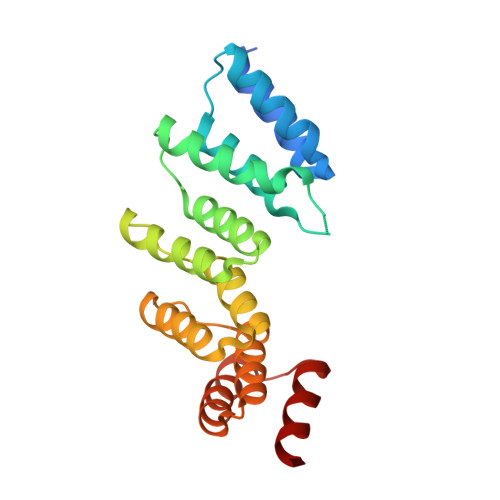
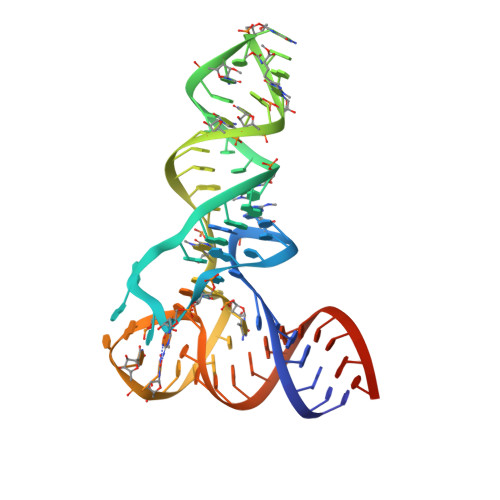
Pentatricopeptide repeats of protein-only RNase P use a distinct mode to recognize conserved bases and structural elements of pre-tRNA.
Teramoto, T., Kaitany, K.J., Kakuta, Y., Kimura, M., Fierke, C.A., Hall, T.M.T.(2020) Nucleic Acids Res 48: 11815-11826
- PubMed: 32719843
- DOI: https://doi.org/10.1093/nar/gkaa627
- Primary Citation of Related Structures:
6LVR - PubMed Abstract:
Pentatricopeptide repeat (PPR) motifs are α-helical structures known for their modular recognition of single-stranded RNA sequences with each motif in a tandem array binding to a single nucleotide. Protein-only RNase P 1 (PRORP1) in Arabidopsis thaliana is an endoribonuclease that uses its PPR domain to recognize precursor tRNAs (pre-tRNAs) as it catalyzes removal of the 5'-leader sequence from pre-tRNAs with its NYN metallonuclease domain. To gain insight into the mechanism by which PRORP1 recognizes tRNA, we determined a crystal structure of the PPR domain in complex with yeast tRNAPhe at 2.85 Å resolution. The PPR domain of PRORP1 bound to the structurally conserved elbow of tRNA and recognized conserved structural features of tRNAs using mechanisms that are different from the established single-stranded RNA recognition mode of PPR motifs. The PRORP1 PPR domain-tRNAPhe structure revealed a conformational change of the PPR domain upon tRNA binding and moreover demonstrated the need for pronounced overall flexibility in the PRORP1 enzyme conformation for substrate recognition and catalysis. The PRORP1 PPR motifs have evolved strategies for protein-tRNA interaction analogous to tRNA recognition by the RNA component of ribonucleoprotein RNase P and other catalytic RNAs, indicating convergence on a common solution for tRNA substrate recognition.
- Epigenetics and Stem Cell Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.
Organizational Affiliation: