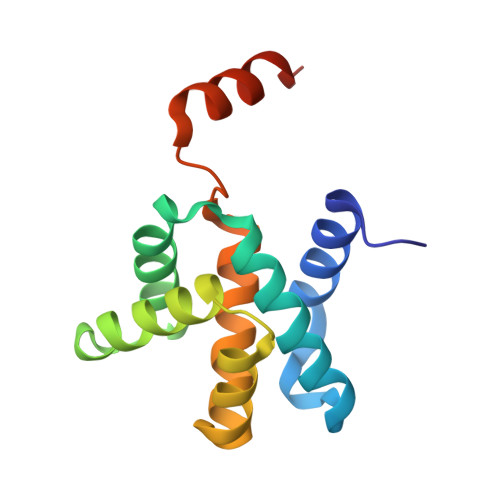
Crystal structure of the Mengla virus VP30 C-terminal domain.
Dong, S., Wen, K., Chu, H., Li, H., Yu, Q., Wang, C., Qin, X.(2020) Biochem Biophys Res Commun 525: 392-397
- PubMed: 32093889
- DOI: https://doi.org/10.1016/j.bbrc.2020.02.089
- Primary Citation of Related Structures:
6LUS - PubMed Abstract:
The family Filoviridae contains many important human viruses, including Marburg virus (MARV) and Ebola virus (EBOV). Měnglà virus (MLAV), a newly discovered filovirus, is considered a potential human pathogen. The VP30 C-terminal domain (CTD) of these filoviruses plays an essential role in virion assembly. In common with other filoviruses, MLAV VP30 CTD mainly exists as a dimer in solution. In this work, we determined the crystal structure of recombinant MLAV VP30 CTD monomer, verifying that C-terminal helix-7 (H7) is critical for the dimerization process. This study provides a preliminary model for investigation of MLAV VP30 CTD as an anti-filovirus drug development target.
- School of Biological Science and Technology, University of Jinan, Jinan, China. Electronic address: bio_dongss@ujn.edu.cn.
Organizational Affiliation: