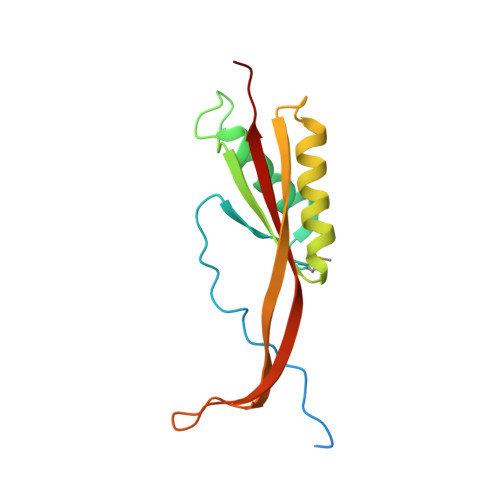
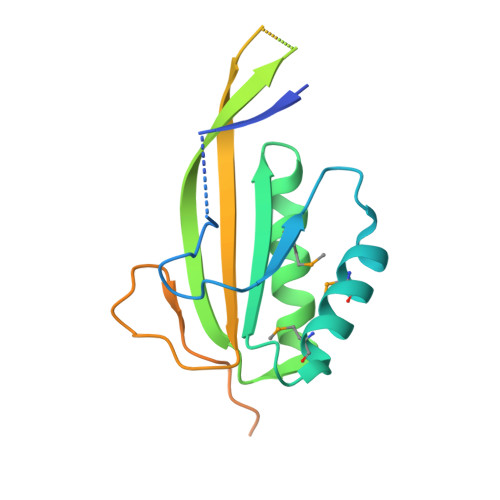
Crystal structure of human RPP20-RPP25 proteins in complex with the P3 domain of lncRNA RMRP.
Yin, C., Bai, G., Zhang, Y., Huang, J.(2021) J Struct Biol 213: 107704-107704
- PubMed: 33571640
- DOI: https://doi.org/10.1016/j.jsb.2021.107704
- Primary Citation of Related Structures:
6LT7 - PubMed Abstract:
Human RNase MRP ribonucleoprotein complex is an essential endoribonuclease involved in the processing of ribosomal RNAs, mitochondrial RNAs and certain messenger RNAs. Its RNA subunit RMRP catalyzes the cleavage of substrate RNAs, and the protein components of RNase MRP are required for activity. RMRP mutations are associated with several types of inherited developmental disorders, but the pathogenic mechanism is largely unknown. Recent structural studies shed lights on the catalytic mechanism of yeast RNase MRP and the closely related RNase P; however, the structural and catalytic mechanism of RMRP in human RNase MRP complex remains unclear. Here we report the crystal structure of the P3 domain of RMRP in complex with the RPP20 and RPP25 proteins of human RNase MRP, which shows that the P3 RNA binds to a conserved positively-charged surface of the RPP20-RPP25 heterodimer through its distal stem and internal loop regions. The disease-related mutations of RMRP P3 are mostly located at the protein-RNA interface and are likely to weaken the binding of P3 to RPP20-RPP25. Moreover, the structure reveals a homodimeric organization of the entire RPP20-RPP25-RMRP P3 complex, which might mediate the dimerization of human RNase MRP complex in cells. These findings provide structural clues to the assembly and pathogenesis of human RNase MRP complex and also reveal a tetrameric feature of RPP20-RPP25 evolutionarily conserved with that of the archaeal Alba proteins.
- State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 201210 Shanghai, China.
Organizational Affiliation: