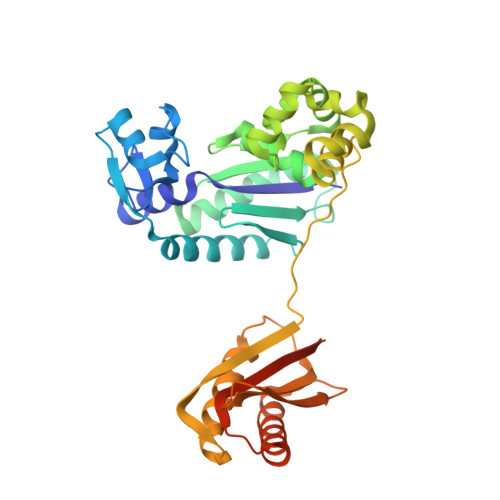
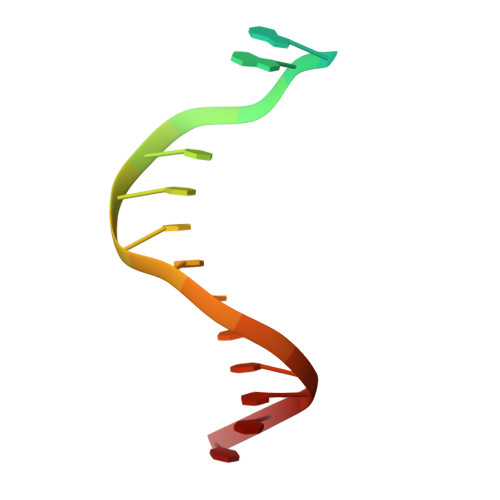
The crystal structure of a natural DNA polymerase complexed with mirror DNA.
An, J., Choi, J., Hwang, D., Park, J., Pemble 4th, C.W., Duong, T.H.M., Kim, K.R., Ahn, H., Chung, H.S., Ahn, D.R.(2020) Chem Commun (Camb) 56: 2186-2189
- PubMed: 31971182
- DOI: https://doi.org/10.1039/c9cc09351f
- Primary Citation of Related Structures:
6L84, 6L97 - PubMed Abstract:
The intrinsic l-DNA binding properties of a natural DNA polymerase was discovered. The binding affinity of Dpo4 polymerase for l-DNA was comparable to that for d-DNA. The crystal structure of Dpo4/l-DNA complex revealed a dimer formed by the little finger domain that provides a binding site for l-DNA.
- Center for Theragnosis, Biomedical Research Institute, Korea Institute of Science and Technology (KIST), Hwarangno 14-gil 5, Seongbuk-gu, Seoul 02792, Republic of Korea. drahn@kist.re.kr and Division of Bio-Medical Science and Technology, KIST School, University of Science and Technology (UST), Seoul 02792, Republic of Korea.
Organizational Affiliation: