Structure Elucidation and Biochemical Characterization of Environmentally Relevant Novel Extradiol Dioxygenases Discovered by a Functional Metagenomics Approach.
Sidhu, C., Solanki, V., Pinnaka, A.K., Thakur, K.G.(2019) mSystems 4
- PubMed: 31771973
- DOI: https://doi.org/10.1128/mSystems.00316-19
- Primary Citation of Related Structures:
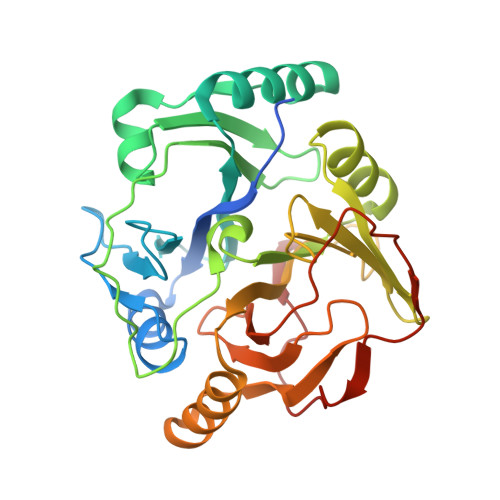
6L3W - PubMed Abstract:
The release of synthetic chemical pollutants in the environment is posing serious health risks. Enzymes, including oxygenases, play a crucial role in xenobiotic degradation. In the present study, we employed a functional metagenomics approach to overcome the limitation of cultivability of microbes under standard laboratory conditions in order to isolate novel dioxygenases capable of degrading recalcitrant pollutants. Fosmid clones possessing dioxygenase activity were further sequenced, and their genes were identified using bioinformatics tools. Two positive fosmid clones, SD3 and RW1, suggested the presence of 2,3-dihydroxybiphenyl 1,2-dioxygenase (BphC-SD3) and catechol 2,3-dioxygenase (C23O-RW1), respectively. Recombinant versions of these enzymes were purified to examine their pollutant-degrading abilities. The crystal structure of BphC-SD3 was determined at 2.6-Å resolution, revealing a two-domain architecture, i.e., N-terminal and C-terminal domains, with the sequential arrangement of βαβββ in each domain, characteristic of Fe-dependent class II type I extradiol dioxygenases. The structure also reveals the presence of conserved amino acids lining the catalytic pocket and Fe 3+ metal ion in the large funnel-shaped active site in the C-terminal domain. Further studies suggest that Fe 3+ bound in the BphC-SD3 active site probably imparts aerobic stability. We further demonstrate the potential application of BphC-SD3 in biosensing of catecholic compounds. The halotolerant and oxygen-resistant properties of these enzymes reported in this study make them potential candidates for bioremediation and biosensing applications. IMPORTANCE The disposal and degradation of xenobiotic compounds have been serious issues due to their recalcitrant properties. Microbial oxygenases are the fundamental enzymes involved in biodegradation that oxidize the substrate by transferring oxygen from molecular oxygen. Among oxygenases, catechol dioxygenases are more versatile in biodegradation and are well studied among the bacterial world. The use of catechol dioxygenases in the field is currently not practical due to their aerobically unstable nature. The significance of our research lies in the discovery of aerobically stable and halotolerant catechol dioxygenases that are efficient in degrading the targeted environmental pollutants and, hence, could be used as cost-effective alternatives for the treatment of hypersaline industrial effluents. Moreover, the structural determination of novel catechol dioxygenases would greatly enhance our knowledge of the function of these enzymes and facilitate directed evolution to further enhance or engineer desired properties.
- Microbial Type Culture Collection and Gene Bank, CSIR-IMTECH, Chandigarh, India.
Organizational Affiliation: