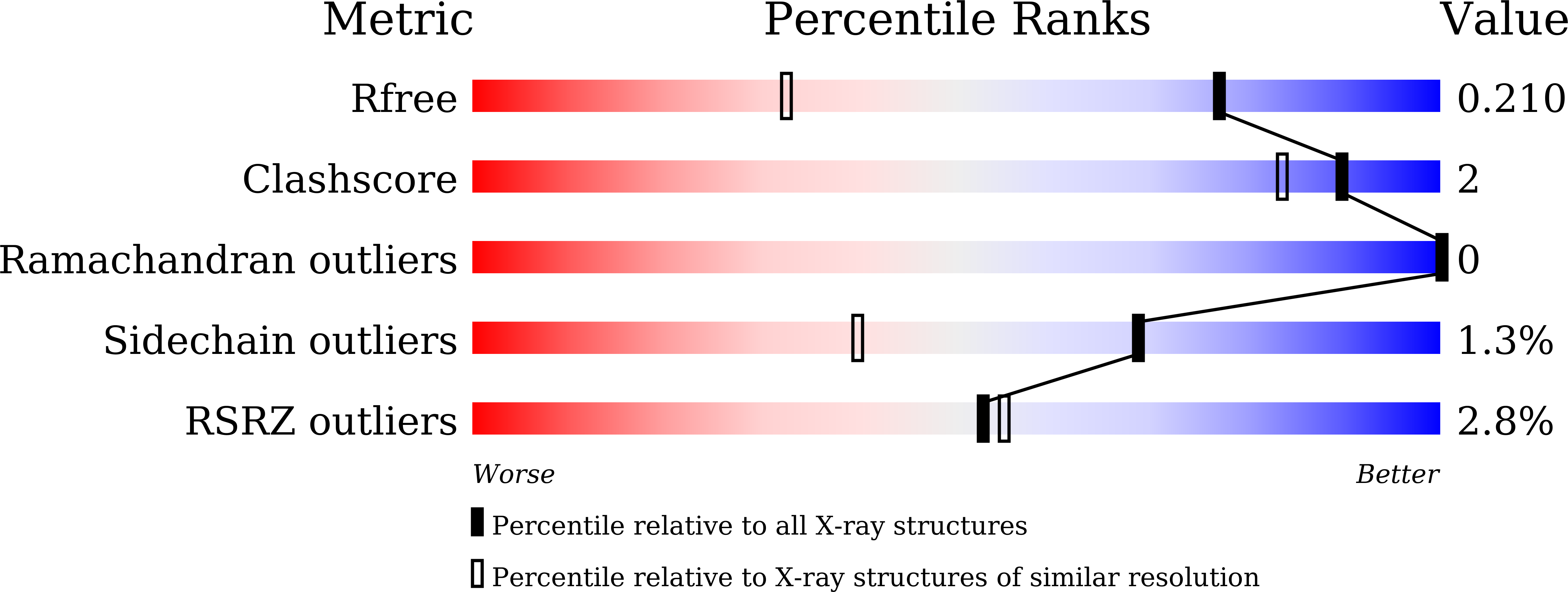
Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Sun, Z., Zhang, H., Zhang, Y., Liao, L., Zhou, W., Zhang, F., Lian, F., Huang, J., Xu, P., Zhang, R., Lu, W., Zhu, M., Tao, H., Yang, F., Ding, H., Chen, S., Yue, L., Zhou, B., Zhang, N., Tan, M., Jiang, H., Chen, K., Liu, B., Liu, C., Dang, Y., Luo, C.(2020) Adv Sci (Weinh) 7: 2000098-2000098
- PubMed: 32714746
- DOI: https://doi.org/10.1002/advs.202000098
- Primary Citation of Related Structures:
6KX2, 6KX3 - PubMed Abstract:
The Rho family GTPases are crucial drivers of tumor growth and metastasis. However, it is difficult to develop GTPases inhibitors due to a lack of well-characterized binding pockets for compounds. Here, through molecular dynamics simulation of the RhoA protein, a groove around cysteine 107 (Cys107) that is relatively well-conserved within the Rho family is discovered. Using a combined strategy, the novel inhibitor DC-Rhoin is discovered, which disrupts interaction of Rho proteins with guanine nucleotide exchange factors (GEFs) and guanine nucleotide dissociation inhibitors (GDIs). Crystallographic studies reveal that the covalent binding of DC-Rhoin to the Cys107 residue stabilizes and captures a novel allosteric pocket. Moreover, the derivative compound DC-Rhoin04 inhibits the migration and invasion of cancer cells, through targeting this allosteric pocket of RhoA. The study reveals a novel allosteric regulatory site within the Rho family, which can be exploited for anti-metastasis drug development, and also provides a novel strategy for inhibitor discovery toward "undruggable" protein targets.
Organizational Affiliation:
School of Life Science and Technology Harbin Institute of Technology Harbin 150001 China.