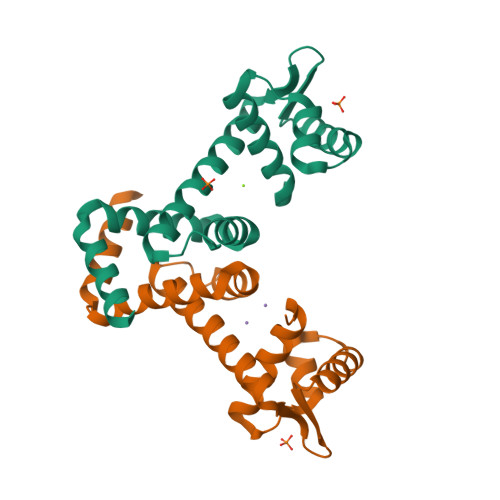
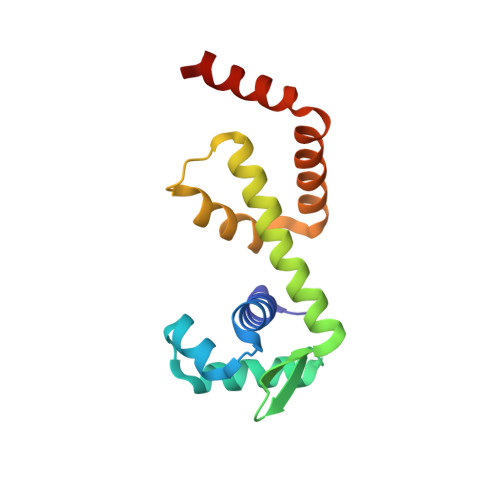
Structural analysis of the manganese transport regulator MntR from Bacillus halodurans in apo and manganese bound forms.
Lee, M.Y., Lee, D.W., Joo, H.K., Jeong, K.H., Lee, J.Y.(2019) PLoS One 14: e0224689-e0224689
- PubMed: 31738781
- DOI: https://doi.org/10.1371/journal.pone.0224689
- Primary Citation of Related Structures:
6KTA, 6KTB - PubMed Abstract:
The manganese transport regulator MntR is a metal-ion activated transcriptional repressor of manganese transporter genes to maintain manganese ion homeostasis. MntR, a member of the diphtheria toxin repressor (DtxR) family of metalloregulators, selectively responds to Mn2+ and Cd2+ over Fe2+, Co2+ and Zn2+. The DtxR/MntR family members are well conserved transcriptional repressors that regulate the expression of metal ion uptake genes by sensing the metal ion concentration. MntR functions as a homo-dimer with one metal ion binding site per subunit. Each MntR subunit contains two domains: an N-terminal DNA binding domain, and a C-terminal dimerization domain. However, it lacks the C-terminal SH3-like domain of DtxR/IdeR. The metal ion binding site of MntR is located at the interface of the two domains, whereas the DtxR/IdeR subunit contains two metal ion binding sites, the primary and ancillary sites, separated by 9 Å. In this paper, we reported the crystal structures of the apo and Mn2+-bound forms of MntR from Bacillus halodurans, and analyze the structural basis of the metal ion binding site. The crystal structure of the Mn2+-bound form is almost identical to the apo form of MntR. In the Mn2+-bound structure, one subunit contains a binuclear cluster of manganese ions, the A and C sites, but the other subunit forms a mononuclear complex. Structural data about MntR from B. halodurans supports the previous hypothesizes about manganese-specific activation mechanism of MntR homologues.
Organizational Affiliation:
Department of Life Science, Dongguk University-Seoul, Ilsandong-gu, Goyang-si, Gyeonggi-do, Republic of Korea.