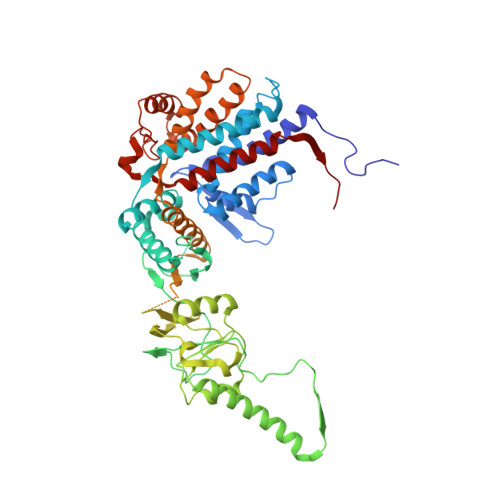
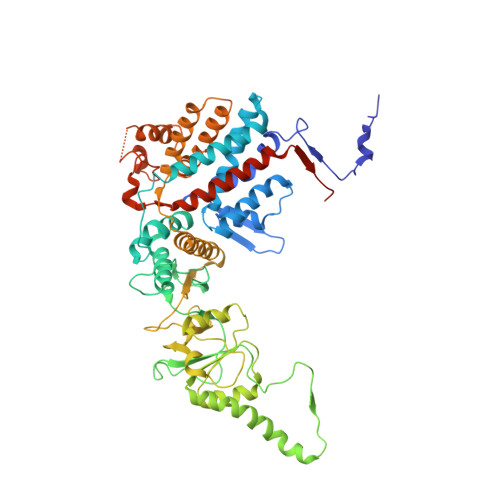
An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Jin, M., Han, W., Liu, C., Zang, Y., Li, J., Wang, F., Wang, Y., Cong, Y.(2019) Proc Natl Acad Sci U S A 116: 19513-19522
- PubMed: 31492816
- DOI: https://doi.org/10.1073/pnas.1903976116
- Primary Citation of Related Structures:
6KRD, 6KRE, 6KS6, 6KS7, 6KS8 - PubMed Abstract:
TRiC/CCT assists the folding of ∼10% of cytosolic proteins through an ATP-driven conformational cycle and is essential in maintaining protein homeostasis. Here, we determined an ensemble of cryo-electron microscopy (cryo-EM) structures of yeast TRiC at various nucleotide concentrations, with 4 open-state maps resolved at near-atomic resolutions, and a closed-state map at atomic resolution, revealing an extra layer of an unforeseen N-terminal allosteric network. We found that, during TRiC ring closure, the CCT7 subunit moves first, responding to nucleotide binding; CCT4 is the last to bind ATP, serving as an ATP sensor; and CCT8 remains ADP-bound and is hardly involved in the ATPase-cycle in our experimental conditions; overall, yeast TRiC consumes nucleotide in a 2-ring positively coordinated manner. Our results depict a thorough picture of the TRiC conformational landscape and its allosteric transitions from the open to closed states in more structural detail and offer insights into TRiC subunit specificity in ATP consumption and ring closure, and potentially in substrate processing.
- National Center for Protein Science Shanghai, State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai 200031, China.
Organizational Affiliation: