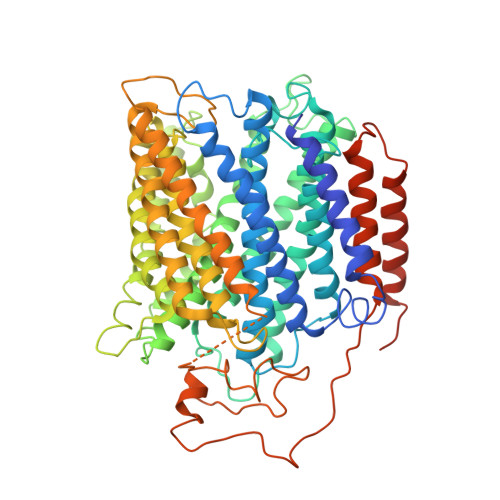
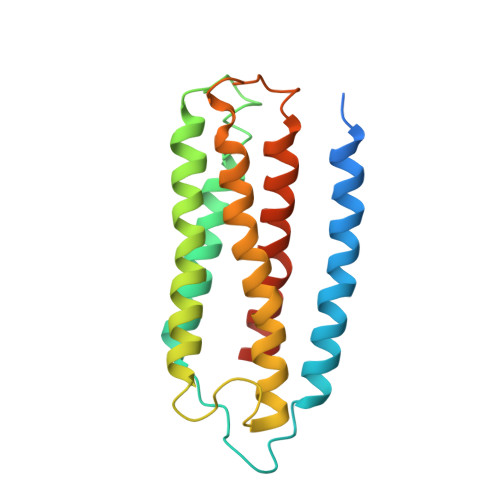
Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Xu, J., Ding, Z., Liu, B., Yi, S.M., Li, J., Zhang, Z., Liu, Y., Li, J., Liu, L., Zhou, A., Gennis, R.B., Zhu, J.(2020) Proc Natl Acad Sci U S A 117: 872-876
- PubMed: 31888984
- DOI: https://doi.org/10.1073/pnas.1915013117
- Primary Citation of Related Structures:
6KOB, 6KOC, 6KOE - PubMed Abstract:
Virtually all proton-pumping terminal respiratory oxygen reductases are members of the heme-copper oxidoreductase superfamily. Most of these enzymes use reduced cytochrome c as a source of electrons, but a group of enzymes have evolved to directly oxidize membrane-bound quinols, usually menaquinol or ubiquinol. All of the quinol oxidases have an additional transmembrane helix (TM0) in subunit I that is not present in the related cytochrome c oxidases. The current work reports the 3.6-Å-resolution X-ray structure of the cytochrome aa 3 -600 menaquinol oxidase from Bacillus subtilis containing 1 equivalent of menaquinone. The structure shows that TM0 forms part of a cleft to accommodate the menaquinol-7 substrate. Crystals which have been soaked with the quinol-analog inhibitor HQNO ( N -oxo-2-heptyl-4-hydroxyquinoline) or 3-iodo-HQNO reveal a single binding site where the inhibitor forms hydrogen bonds to amino acid residues shown previously by spectroscopic methods to interact with the semiquinone state of menaquinone, a catalytic intermediate.
- School of Medicine and Life Sciences, Nanjing University of Chinese Medicine, 210023 Nanjing, China.
Organizational Affiliation: