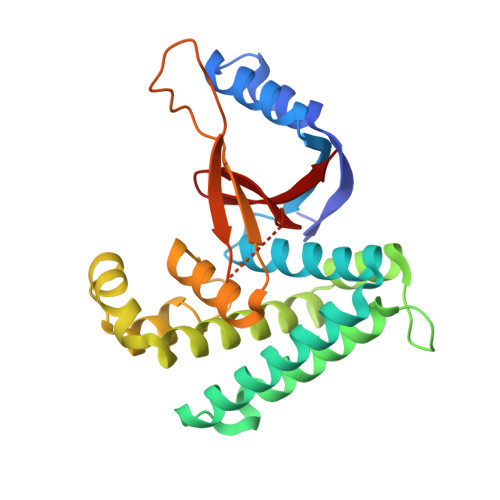
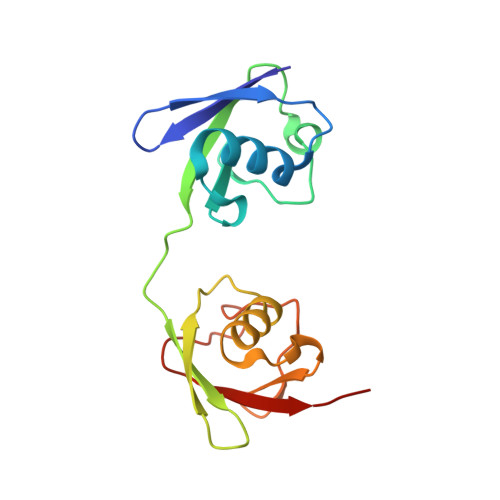
Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Lu, L., Zhai, X., Li, X., Wang, S., Zhang, L., Wang, L., Jin, X., Liang, L., Deng, Z., Li, Z., Wang, Y., Fu, X., Hu, H., Wang, J., Mei, Z., He, Z., Wang, F.(2022) Nat Commun 13: 4672