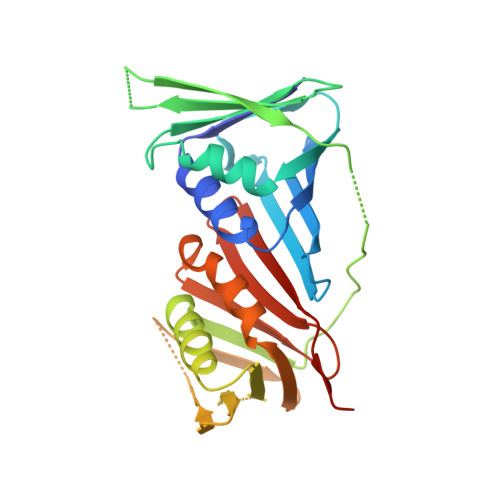
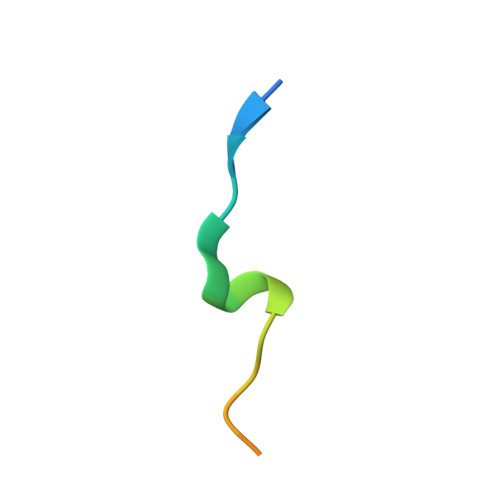
Structure of PCNA in complex with DNMT1 PIP box reveals the basis for the molecular mechanism of the interaction.
Jimenji, T., Matsumura, R., Kori, S., Arita, K.(2019) Biochem Biophys Res Commun 516: 578-583
- PubMed: 31235252
- DOI: https://doi.org/10.1016/j.bbrc.2019.06.060
- Primary Citation of Related Structures:
6K3A - PubMed Abstract:
DNMT1 is a C5-DNA methyltransferase that plays a pivotal role in DNA methylation maintenance. During early and mid S-phase, DNMT1 accumulates at DNA replication sites by binding to proliferating cell nuclear antigen (PCNA), an essential factor for DNA replication, through a PIP box motif. However, the molecular mechanism by which the DNMT1 PIP box motif binds to PCNA remains unclear. Here, we report the crystal structure of PCNA bound to DNMT1 PIP box peptide. The structure reveals the detailed interaction between PCNA and DNMT1 PIP box; conserved glutamine and hydrophobic/aromatic residues in the PIP box are recognized by the Q- and hydrophobic pockets of PCNA, respectively. The structure also shows novel intramolecular interactions within the PIP box motif, which stabilize the helix conformation in the PIP box. Our data provide structural insight into the recruitment of DNMT1 to replication sites by PCNA.
- Structural Biology Laboratory, Graduate School of Medical Life Science, Yokohama City University, Yokohama, 230-0045, Japan.
Organizational Affiliation: