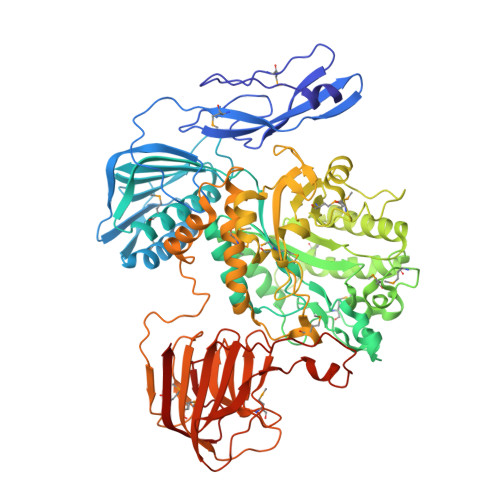
Biochemical characteristics and crystallographic evidence for substrate-assisted catalysis of a beta-N-acetylhexosaminidase in Akkermansia muciniphila.
Chen, X., Li, M., Wang, Y., Tang, R., Zhang, M.(2019) Biochem Biophys Res Commun 517: 29-35
- PubMed: 31345574
- DOI: https://doi.org/10.1016/j.bbrc.2019.06.150
- Primary Citation of Related Structures:
6JQF - PubMed Abstract:
In this paper, we characterized Am2136 as a β-N-acetylhexosaminidase from Akkermansia muciniphila to perform the biochemical characteristics and the crystal structure of selenomethionine-labeled Am2136 with GlcNAc complex. Crystallographic evidence suggests that an oxazolinium ion was formed intermediately by the 2-acetamido group during the substrate-assisted catalytic procedure. Structural and kinetic analysis of native Am2136 and D412A, E413A mutants were investigated and the results revealed substantial difference. The K cat /K m value of D412A was decreased 4297-fold compared to native Am2136 revealed that mutation of Asp-412 results in preventing the 2-acetamido substituent from providing anchimeric assistance and thus reducing the catalytic efficiency. Moreover, Am2136 has a wide dependence on pH and temperature, while sensitive to divalent metal ions such as Ca 2+ and Mn 2+ . These biochemical and crystallographic results provide evidences that Asp-412 residue assists to orient the 2-acetamido group for catalysis. Based on crystallographic evidence and sequence alignment with other GH family 20 enzymes, Asp-412 residue is possibly fundamental for Am2136 during substrate-assisted catalysis.
- School of Life Sciences, Anhui University, 111 Jiulong Road, Hefei, Anhui, China; Key Laboratory of Human Microenvironment and Precision Medicine of Anhui Higher Education Institute, Anhui University, 111 Jiulong Road, Hefei, Anhui, China.
Organizational Affiliation: