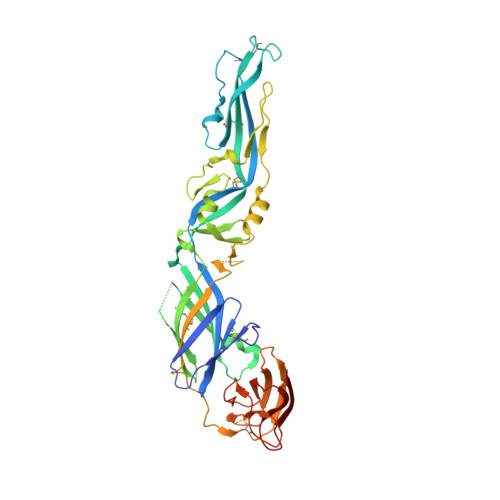
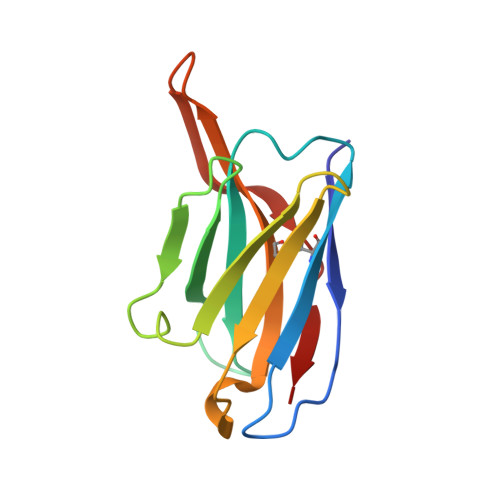
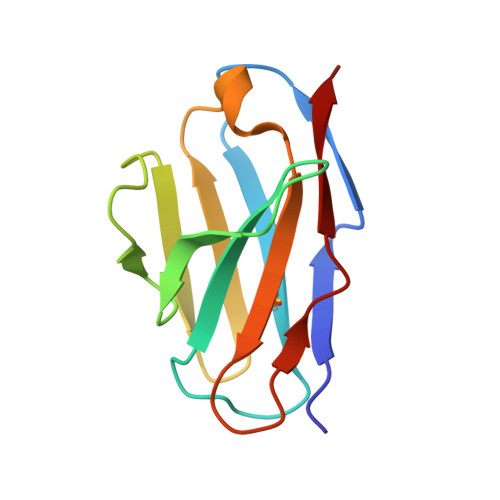
Double Lock of a Human Neutralizing and Protective Monoclonal Antibody Targeting the Yellow Fever Virus Envelope.
Lu, X., Xiao, H., Li, S., Pang, X., Song, J., Liu, S., Cheng, H., Li, Y., Wang, X., Huang, C., Guo, T., Ter Meulen, J., Daffis, S., Yan, J., Dai, L., Rao, Z., Klenk, H.D., Qi, J., Shi, Y., Gao, G.F.(2019) Cell Rep 26: 438-446.e5
- PubMed: 30625326
- DOI: https://doi.org/10.1016/j.celrep.2018.12.065
- Primary Citation of Related Structures:
6IVZ, 6IW0, 6IW1, 6IW2, 6IW4, 6IW5 - PubMed Abstract:
Yellow fever virus (YFV), a deadly human pathogen, is the prototype of the genus Flavivirus. Recently, YFV re-emerged in Africa and Brazil, leading to hundreds of deaths, with some cases imported to China. Prophylactic or therapeutic countermeasures are urgently needed. Previously, several human monoclonal antibodies against YFV were screened out by phage display. Here, we find that one of them, 5A, exhibits high neutralizing potency and good protection. Crystallographic analysis of the YFV envelope (E) protein in its pre- and post-fusion states shows conformations similar to those observed in other E proteins of flaviviruses. Furthermore, the structures of 5A in complex with the E protein in both states are resolved, revealing an invariant recognition site. Structural analysis and functional data suggest that 5A has high neutralization potency because it interferes with virus entry by preventing both virus attachment and fusion. These findings will be instrumental for immunogen or inhibitor design.
- Laboratory of Protein Engineering and Vaccines, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China.
Organizational Affiliation: