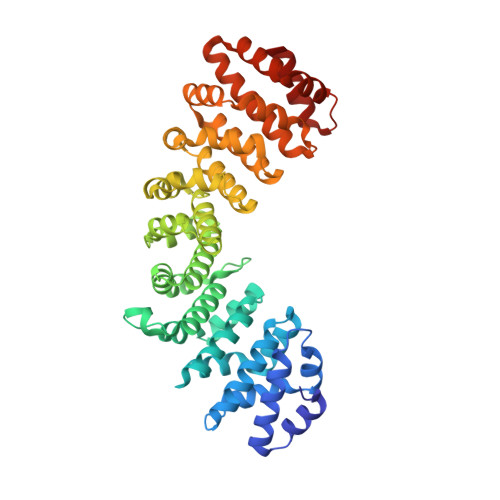
Structural and biochemical characterization of the recognition of the 53BP1 nuclear localization signal by importin-alpha.
Matsuura, Y.(2019) Biochem Biophys Res Commun 510: 236-241
- PubMed: 30685087
- DOI: https://doi.org/10.1016/j.bbrc.2019.01.075
- Primary Citation of Related Structures:
6IU7, 6IUA - PubMed Abstract:
53BP1 (TP53-binding protein 1) plays a key role in DNA double-strand break repair by promoting non-homologous end joining (NHEJ) especially during G1 phase of the cell cycle. Nuclear import of 53BP1 is required for proper localization of 53BP1 and maintenance of genome integrity. 53BP1 has a classical bipartite nuclear localization signal (NLS) of sequence 1666-GKRKLITSEEERSPAKRGRKS-1686. Ser1678 within the 53BP1 NLS can be phosphorylated by CDK1/cyclin B, and a phosphomimetic substitution of Ser1678 with aspartate has been shown to negatively regulate nuclear import of 53BP1. Here, the X-ray crystal structures of the nuclear import adaptor importin-α1 bound to the wild-type 53BP1 NLS and the S1678D mutant of 53BP1 NLS are reported at resolutions of 1.9 and 1.7 Å, respectively. In the wild-type structure, not only the two basic clusters of the 53BP1 NLS but also the linker region between the basic clusters made extensive interactions with importin-α1. In the mutant structure, the linker region between the basic clusters in the 53BP1 NLS made fewer interactions with importin-α1 than those observed in the wild-type structure. However, biochemical binding assays using purified proteins showed that the 53BP1 mutation S1678D reduces the binding affinity to importin-α1 only to a modest extent. Implications of these findings for regulatory mechanism of 53BP1 nuclear import are discussed.
- Division of Biological Science, Graduate School of Science, Nagoya University, Japan; Structural Biology Research Center, Graduate School of Science, Nagoya University, Japan. Electronic address: matsuura.yoshiyuki@d.mbox.nagoya-u.ac.jp.
Organizational Affiliation: