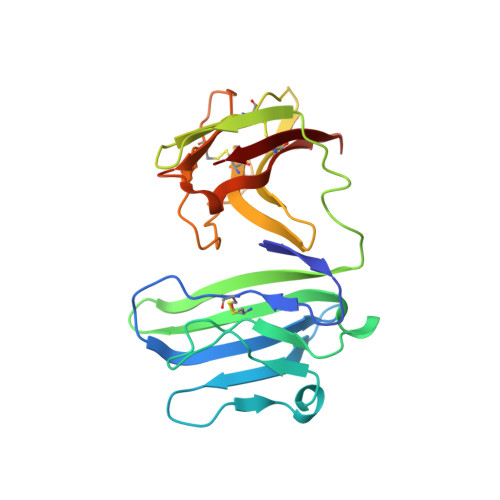
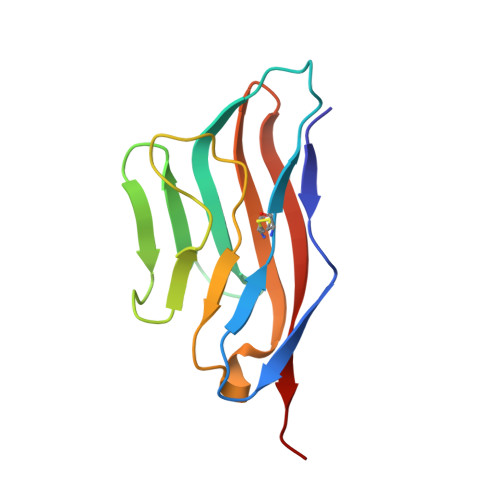
Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Wang, H., Qi, J., Zhang, S., Li, Y., Tan, S., Gao, G.F.(2019) Proc Natl Acad Sci U S A 116: 988-996
- PubMed: 30591568
- DOI: https://doi.org/10.1073/pnas.1815716116
- Primary Citation of Related Structures:
6ISA, 6ISB, 6ISC - PubMed Abstract:
Natural killer (NK) cells are important component of innate immunity and also contribute to activating and reshaping the adaptive immune responses. The functions of NK cells are modulated by multiple inhibitory and stimulatory receptors. Among these receptors, the activating receptor CD226 (DNAM-1) mediates NK cell activation via binding to its nectin-like (Necl) family ligand, CD155 (Necl-5). Here, we present a unique side-by-side arrangement pattern of two tandem immunoglobulin V-set (IgV) domains deriving from the ectodomains of both human CD226 (hCD226-ecto) and mouse CD226 (mCD226-ecto), which is substantially different from the conventional head-to-tail arrangement of other multiple Ig-like domain molecules. The hybrid complex structure of mCD226-ecto binding to the first domain of human CD155 (hCD155-D1) reveals a conserved binding interface with the first domain of CD226 (D1), whereas the second domain of CD226 (D2) both provides structural supports for the unique architecture of CD226 and forms direct interactions with CD155. In the absence of the D2 domain, CD226-D1 exhibited substantially reduced binding efficacy to CD155. Collectively, these findings would broaden our knowledge of the interaction between NK cell receptors and the nectin/Necl family ligands, as well as provide molecular basis for the development of CD226-targeted antitumor immunotherapeutics.
- Research Network of Immunity and Health (RNIH), Beijing Institutes of Life Science, Chinese Academy of Sciences (CAS), 100101 Beijing, China.
Organizational Affiliation: