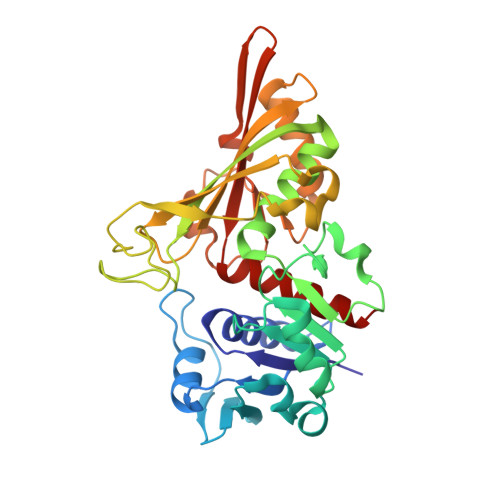
Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase from Lactobacillus plantarum: Insight into the Mercury Binding Mechanism
Yoneda, K., Ogata, M., Nishiyama, K., Fukuda, K., Yasuda, S., Igoshi, K., Kinoshita, H.(2019) Milk Sci 68: 3-11