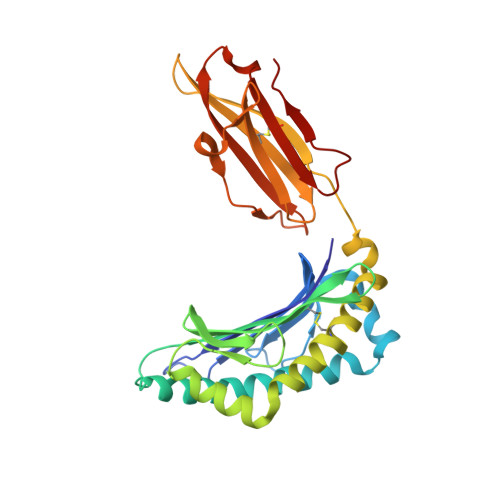
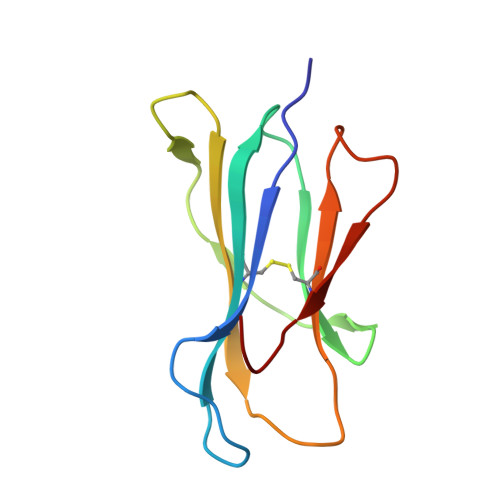
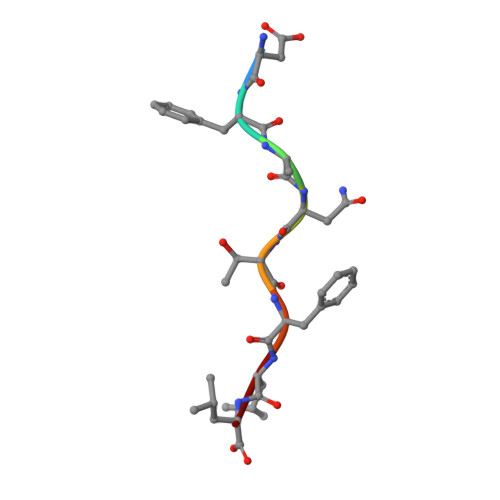
Structure and Peptidome of the Bat MHC Class I Molecule Reveal a Novel Mechanism Leading to High-Affinity Peptide Binding.
Qu, Z., Li, Z., Ma, L., Wei, X., Zhang, L., Liang, R., Meng, G., Zhang, N., Xia, C.(2019) J Immunol 202: 3493-3506
- PubMed: 31076531
- DOI: https://doi.org/10.4049/jimmunol.1900001
- Primary Citation of Related Structures:
6ILC, 6ILE, 6ILF, 6ILG - PubMed Abstract:
Bats are natural reservoir hosts, harboring more than 100 viruses, some of which are lethal to humans. The asymptomatic coexistence with viruses is thought to be connected to the unique immune system of bats. MHC class I (MHC I) presentation is closely related to cytotoxic lymphocyte immunity, which plays an important role in viral resistance. To investigate the characteristics of MHC I presentation in bats, the crystal structures of peptide-MHC I complexes of Pteropus alecto , Ptal-N*01:01/HEV-1 (DFANTFLP) and Ptal-N*01:01/HEV-2 (DYINTNLVP), and two related mutants, Ptal-N*01:01/HEV-1 PΩL (DFANTFLL) and Ptal-N*01:01 ΔMDL /HEV-1, were determined. Through structural analysis, we found that Ptal-N*01:01 had a multi-Ala-assembled pocket B and a flexible hydrophobic pocket F, which could accommodate variable anchor residues and allow Ptal-N*01:01 to bind numerous peptides. Three sequential amino acids, Met, Asp, and Leu, absent from the α1 domain of the H chain in other mammals, were present in this domain in the bat. Upon deleting these amino acids and determining the structure in p/Ptal-N*01:01 ΔMDL /HEV-1, we found they helped form an extra salt-bridge chain between the H chain and the N-terminal aspartic acid of the peptide. By introducing an MHC I random peptide library for de novo liquid chromatography-tandem mass spectrometry analysis, we found that this insertion module, present in all types of bats, can promote MHC I presentation of peptides with high affinity during the peptide exchange process. This study will help us better understand how bat MHC I presents high-affinity peptides from an extensive binding peptidome and provides a foundation to understand the cellular immunity of bats.
- Department of Microbiology and Immunology, College of Veterinary Medicine, China Agricultural University, Haidian District, Beijing 100094, China.
Organizational Affiliation: