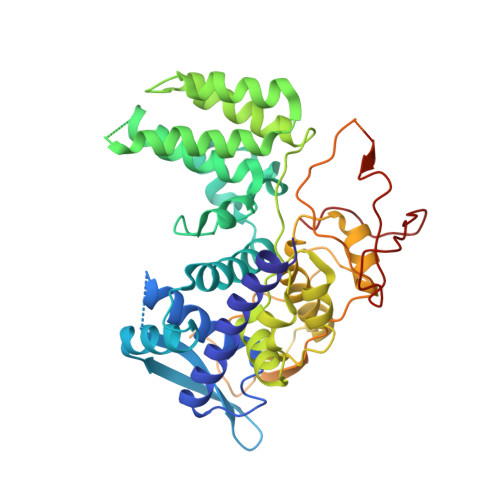
Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Coloma, R., Arranz, R., de la Rosa-Trevin, J.M., Sorzano, C.O.S., Munier, S., Carlero, D., Naffakh, N., Ortin, J., Martin-Benito, J.(2020) Nat Microbiol 5: 727-734
- PubMed: 32152587
- DOI: https://doi.org/10.1038/s41564-020-0675-3
- Primary Citation of Related Structures:
6H9G, 6I54, 6I7B, 6I7M, 6I85 - PubMed Abstract:
The influenza virus genome consists of eight viral ribonucleoproteins (vRNPs), each consisting of a copy of the polymerase, one of the genomic RNA segments and multiple copies of the nucleoprotein arranged in a double helical conformation. vRNPs are macromolecular machines responsible for messenger RNA synthesis and genome replication, that is, the formation of progeny vRNPs. Here, we describe the structural basis of the transcription process. The mechanism, which we call the 'processive helical track', is based on the extreme flexibility of the helical part of the vRNP that permits a sliding movement between both antiparallel nucleoprotein-RNA strands, thereby allowing the polymerase to move over the genome while bound to both RNA ends. Accordingly, we demonstrate that blocking this movement leads to inhibition of vRNP transcriptional activity. This mechanism also reveals a critical role of the nucleoprotein in maintaining the double helical structure throughout the copying process to make the RNA template accessible to the polymerase.
- Centro Nacional de Biotecnología, Consejo Superior de Investigaciones Científicas (CSIC), Madrid, Spain.
Organizational Affiliation: