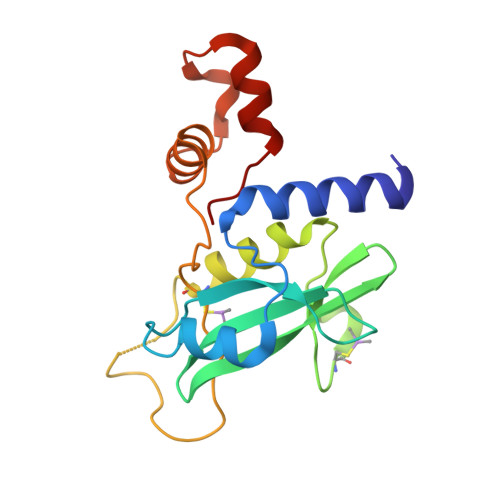
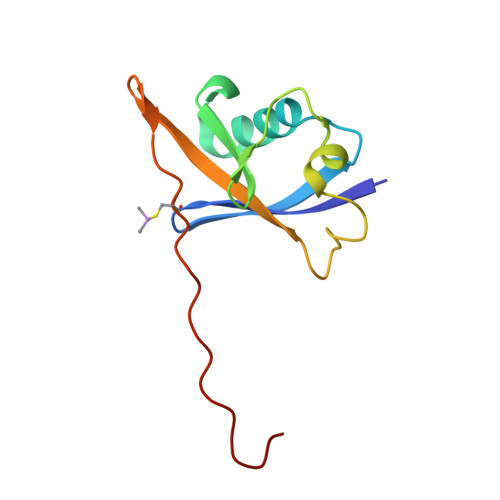
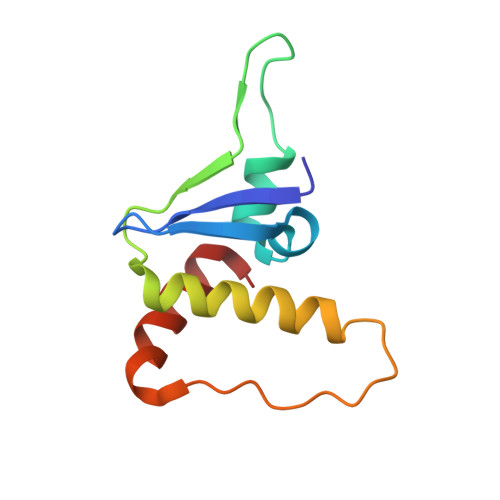
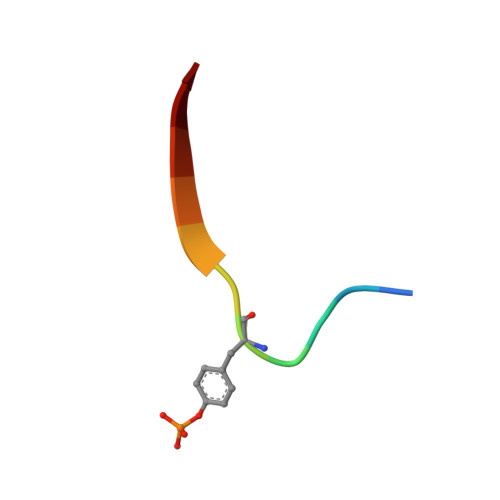
Structural insights into substrate recognition by the SOCS2 E3 ubiquitin ligase.
Kung, W.W., Ramachandran, S., Makukhin, N., Bruno, E., Ciulli, A.(2019) Nat Commun 10: 2534-2534
- PubMed: 31182716
- DOI: https://doi.org/10.1038/s41467-019-10190-4
- Primary Citation of Related Structures:
6I4X, 6I5J, 6I5N - PubMed Abstract:
The suppressor of cytokine signaling 2 (SOCS2) acts as substrate recognition subunit of a Cullin5 E3 ubiquitin ligase complex. SOCS2 binds to phosphotyrosine-modified epitopes as degrons for ubiquitination and proteasomal degradation, yet the molecular basis of substrate recognition has remained elusive. Here, we report co-crystal structures of SOCS2-ElonginB-ElonginC in complex with phosphorylated peptides from substrates growth hormone receptor (GHR-pY595) and erythropoietin receptor (EpoR-pY426) at 1.98 Å and 2.69 Å, respectively. Both peptides bind in an extended conformation recapitulating the canonical SH2 domain-pY pose, but capture different conformations of the EF loop via specific hydrophobic interactions. The flexible BG loop is fully defined in the electron density, and does not contact the substrate degron directly. Cancer-associated SNPs located around the pY pocket weaken substrate-binding affinity in biophysical assays. Our findings reveal insights into substrate recognition and specificity by SOCS2, and provide a blueprint for small molecule ligand design.
- Division of Biological Chemistry and Drug Discovery, School of Life Sciences, University of Dundee, James Black Centre, Dow Street, Dundee, DD1 5EH, UK.
Organizational Affiliation: