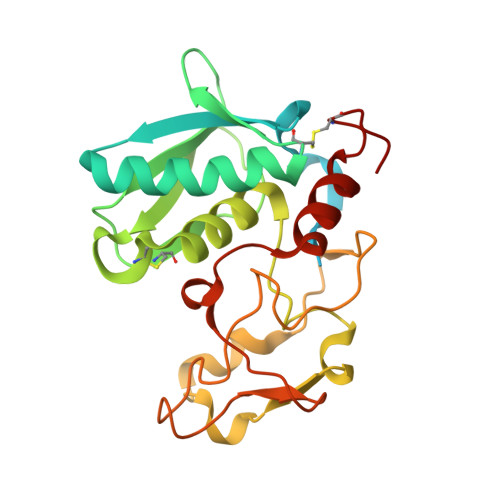
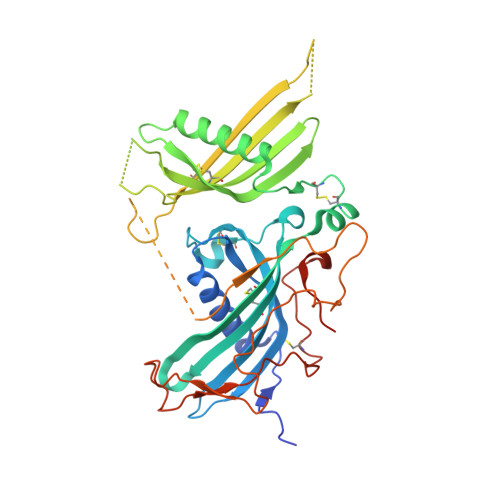
Structure of mammalian plasma fetuin-B and its mechanism of selective metallopeptidase inhibition.
Cuppari, A., Korschgen, H., Fahrenkamp, D., Schmitz, C., Guevara, T., Karmilin, K., Kuske, M., Olf, M., Dietzel, E., Yiallouros, I., de Sanctis, D., Goulas, T., Weiskirchen, R., Jahnen-Dechent, W., Floehr, J., Stoecker, W., Jovine, L., Gomis-Ruth, F.X.(2019) IUCrJ 6: 317-330
- PubMed: 30867929
- DOI: https://doi.org/10.1107/S2052252519001568
- Primary Citation of Related Structures:
6HPV, 6HT9 - PubMed Abstract:
Mammalian fetuin-A and fetuin-B are abundant serum proteins with pleiotropic functions. Fetuin-B is a highly selective and potent inhibitor of metallo-peptidases (MPs) of the astacin family, which includes ovastacin in mammals. By inhibiting ovastacin, fetuin-B is essential for female fertility. The crystal structure of fetuin-B was determined unbound and in complex with archetypal astacin, and it was found that the inhibitor has tandem cystatin-type modules (CY1 and CY2). They are connected by an exposed linker with a rigid, disulfide-linked 'CPDCP-trunk', and are followed by a C-terminal region (CTR) with little regular secondary structure. The CPDCP-trunk and a hairpin of CY2 form a bipartite wedge, which slots into the active-site cleft of the MP. These elements occupy the nonprimed and primed sides of the cleft, respectively, but spare the specificity pocket so that the inhibitor is not cleaved. The aspartate in the trunk blocks the catalytic zinc of astacin, while the CY2 hairpin binds through a QWV X GP motif. The CY1 module assists in structural integrity and the CTR is not involved in inhibition, as verified by in vitro studies using a cohort of mutants and variants. Overall, the inhibition conforms to a novel 'raised-elephant-trunk' mechanism for MPs, which is reminiscent of single-domain cystatins that target cysteine peptidases. Over 200 sequences from vertebrates have been annotated as fetuin-B, underpinning its ubiquity and physiological relevance; accordingly, sequences with conserved CPDCP- and QWV X GP-derived motifs have been found from mammals to cartilaginous fishes. Thus, the raised-elephant-trunk mechanism is likely to be generally valid for the inhibition of astacins by orthologs of fetuin-B.
- Proteolysis Laboratory, Department of Structural Biology, Molecular Biology Institute of Barcelona, CSIC, Barcelona Science Park, Helix Building, c/o Baldiri Reixac 15-21, E-08028 Barcelona, Catalonia, Spain.
Organizational Affiliation: