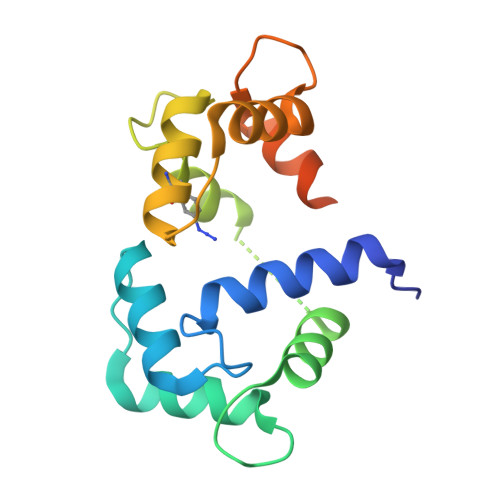
Conformation-specific detection of calmodulin binding using the unnatural amino acid p-azido-phenylalanine (AzF) as an IR-sensor.
Creon, A., Josts, I., Niebling, S., Huse, N., Tidow, H.(2018) Struct Dyn 5: 064701-064701
- PubMed: 30474048
- DOI: https://doi.org/10.1063/1.5053466
- Primary Citation of Related Structures:
6HCS - PubMed Abstract:
Calmodulin (CaM) is a very conserved, ubiquitous, eukaryotic protein that binds four Ca 2+ ions with high affinity. It acts as a calcium sensor by translating Ca 2+ signals into cellular processes such as metabolism, inflammation, immune response, memory, and muscle contraction. Calcium binding to CaM leads to conformational changes that enable Ca 2+ /CaM to recognize and bind various target proteins with high affinity. The binding mode and binding partners of CaM are very diverse, and a consensus binding sequence is lacking. Here, we describe an elegant system that allows conformation-specific detection of CaM-binding to its binding partners. We incorporate the unnatural amino acid p-azido-phenylalanine (AzF) in different positions of CaM and follow its unique spectral signature by infrared (IR)-spectroscopy of the azido stretching vibration. Our results suggest that the AzF vibrational probe is sensitive to the chemical environment in different CaM/CaM-binding domain (CaMBD) complexes, which allows differentiating between different binding motifs according to the spectral characteristics of the azido stretching mode. We corroborate our results with a crystal structure of AzF-labelled CaM (CaM108AzF) in complex with a binding peptide from calmodulin-dependent protein kinase IIα identifying the structural basis for the observed IR frequency shifts.