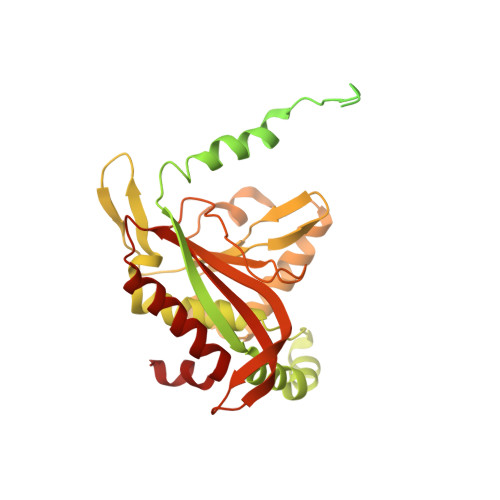
Structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate dehydrogenase complex (hKGDHc) revealed by cryo-EM and cross-linking mass spectrometry: Implications for the overall hKGDHc structure.
Nagy, B., Polak, M., Ozohanics, O., Zambo, Z., Szabo, E., Hubert, A., Jordan, F., Novacek, J., Adam-Vizi, V., Ambrus, A.(2021) Biochim Biophys Acta Gen Subj 1865: 129889-129889
- PubMed: 33684457
- DOI: https://doi.org/10.1016/j.bbagen.2021.129889
- Primary Citation of Related Structures:
6H05 - PubMed Abstract:
The human mitochondrial alpha-ketoglutarate dehydrogenase complex (hKGDHc) converts KG to succinyl-CoA and NADH. Malfunction of and reactive oxygen species generation by the hKGDHc as well as its E1-E2 subcomplex are implicated in neurodegenerative disorders, ischemia-reperfusion injury, E3-deficiency and cancers. We performed cryo-EM, cross-linking mass spectrometry (CL-MS) and molecular modeling analyses to determine the structure of the E2 component of the hKGDHc (hE2k); hE2k transfers a succinyl group to CoA and forms the structural core of hKGDHc. We also assessed the overall structure of the hKGDHc by negative-stain EM and modeling. We report the 2.9 Å resolution cryo-EM structure of the hE2k component. The cryo-EM map comprises density for hE2k residues 151-386 - the entire (inner) core catalytic domain plus a few additional residues -, while residues 1-150 are not observed due to the inherent flexibility of the N-terminal region. The structure of the latter segment was also determined by CL-MS and homology modeling. Negative-stain EM on in vitro assembled hKGDHc and previous data were used to build a putative overall structural model of the hKGDHc. The E2 core of the hKGDHc is composed of 24 hE2k chains organized in octahedral (8 × 3 type) assembly. Each lipoyl domain is oriented towards the core domain of an adjacent chain in the hE2k homotrimer. hE1k and hE3 are most likely tethered at the edges and faces, respectively, of the cubic hE2k assembly. The revealed structural information will support the future pharmacologically targeting of the hKGDHc.
- Department of Biochemistry, Institute of Biochemistry and Molecular Biology, Semmelweis University, Budapest, Hungary.
Organizational Affiliation: